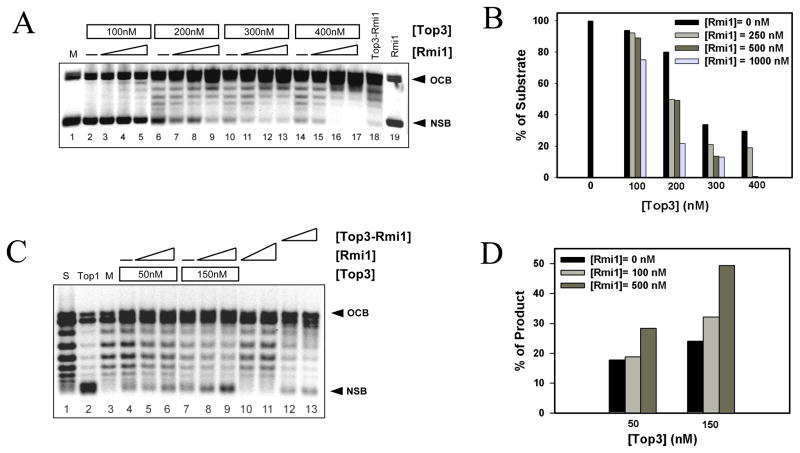
Fig. 3.
Rmi1 stimulates Top3-dependent relaxation of supercoiled bubble DNA at 30°C. (A) Negatively supercoiled DNA containing a 500 bp bubble (NSB) was prepared as described (30) and incubated with the indicated concentrations of Top3 together with increasing concentrations of Rmi1 (0, 250, 500, or 1000 nM) for 20 min at 30°C. Alternatively, NSB was mock treated (lane 1), or treated with 125 nM Top3-Rmi1 (lane 18) or 1000 nM Rmi1 (lane 19). OCB, Open circular bubble DNA. The products were analyzed by native gel electrophoresis as in Fig 2A. (B) The percent of initial substrate DNA (NSB; mock = 100%) remaining in each reaction was quantified and is presented as a function of Top3 concentration. (C) Positively supercoiled DNA containing a 500 bp bubble (PSB) was prepared by pre-incubating OCB with EtBr. This substrate was then incubated with the indicated concentration of Top3 together with increasing amounts of Rmi1 (0, 100, 500 nM) for 20 min at 30°C. Alternatively, PSB substrate (S, lane 1) was treated with DNA topoisomerase I (Top1, lane 2), mock treated (M, lane 3), or treated with 50 nM (lane 12) or 100 nM (lane 13) Top3-Rmi1. Following incubation, the DNA was purified away from protein and EtBr by phenol/chloroform extraction and analyzed by native agarose gel electrophoresis. Note that the relaxation of PSB DNA produces NSB DNA. (D) The NSB product DNA was quantified for each reaction and is presented as percent maximal relaxation (where M = 0% and Top1 = 100%).