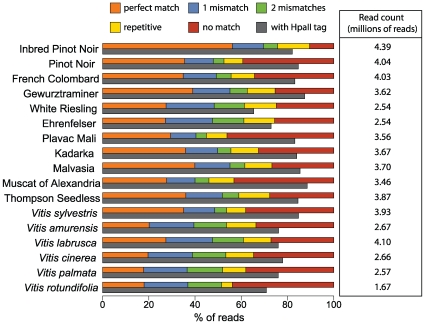
Figure 1. Alignment results of Illumina GA reads to the grapevine reference genome.
The total number of reads generated for each sample is found in the box to the right. The upper bars in the barplot indicate the proportion of reads belonging to each of the categories in the legend. Reads aligning with 0 to 2 mismatches were included for SNP discovery. Reads mapped repetitively and reads with no match were discarded. The lower bars (dark grey) show the proportion of reads beginning with the HpaII tag. The wild Vitis samples are shown in italics.