Figure 5.
Activation-Tagged and Loss-of-Function Alleles of AP2-Like Target Genes.
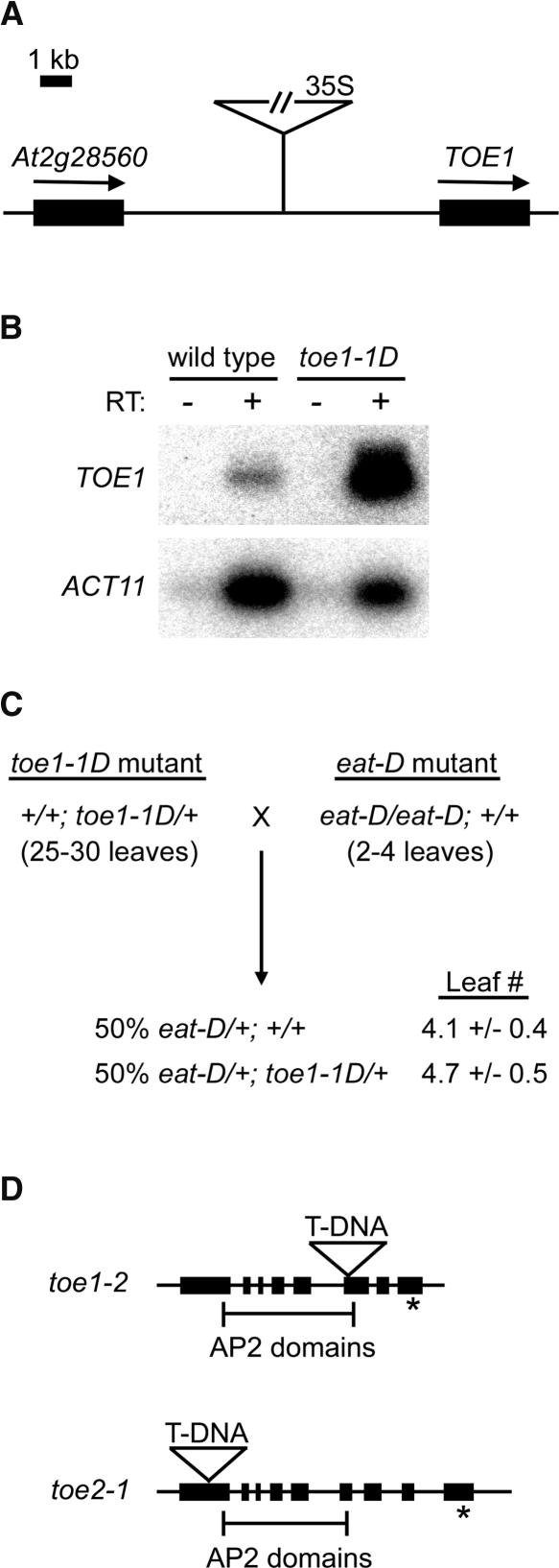
(A) Location of the T-DNA insert in toe1-1D. The 35S enhancers are located ∼5 kb upstream of TOE1 (At2g28550).
(B) Semiquantitative RT-PCR analysis of TOE1 and ACT11 expression in the wild type versus toe1-1D. RNA from each genotype was isolated from floral buds. Inclusion of reverse transcriptase (RT) is indicated by + or − above the gels.
(C) eat-D suppresses the phenotype of toe1-1D. Flowering-time phenotypes (expressed as rosette leaf number) of F1 plants derived from the cross of a toe1-1D heterozygote to an eat-D homozygote. Approximately 50% of the F1 plants contained the toe1-1D allele, based on PCR genotyping; these plants were early flowering, indicating the suppression of toe1-1D (which overexpresses TOE1) by eat-D (which overexpresses miR172). In this experiment, wild-type control plants flowered with ∼11 leaves.
(D) Locations of the T-DNA inserts in toe1-2 and toe2-1. Exons are indicated by solid bars, and the target sequences for miR172 are indicated with asterisks. Brackets delineate the two AP2 domains contained in each gene.