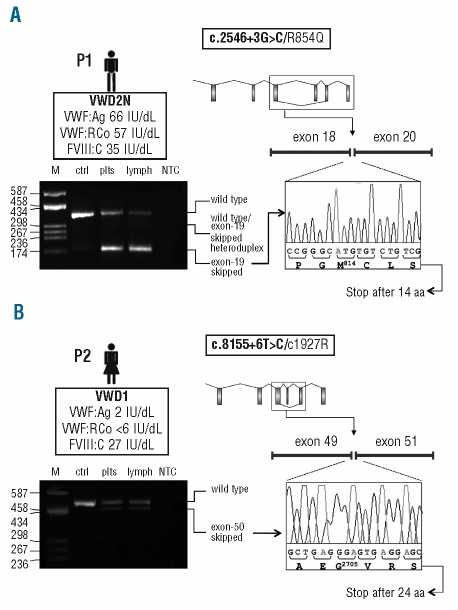
Figure 1.
Functional analysis of the effect of the c.2546+3G>C and c.8155+6T>C splicing mutations on the VWF mRNA. (A, B) Top left: In the box underneath the patient’s ideogram: patient’s VWD type (in bold), VWF:Ag, VWF:RCo, and FVIII:C levels are listed. Top right: Schematic representation of the VWF gene. Exons (boxes) and introns (lines) are drawn to scale. The aberrant splicing event observed in each patient (i.e. exon-19 and exon-50 skipping for patients carrying the c.2546+3G>C or the c.8155+6T>C mutations, respectively) is represented. The patient’s mutations are indicated in the box above the gene scheme, with the splicing mutation in bold. Mutation numbering is according to GenBank accession numbers NM_000552.3 (according to cDNA position) and NP_000543.2 (native protein). Bottom left: RT-PCR products separated on a 2% agarose gel. Lane M: molecular weight marker (pUC8-HaeIII); other lanes (from left to right): RT-PCR products amplified from total RNA extracted from patients’ platelets (plts) and lymphocytes (lymph), and from platelet-derived RNA of a wild-type individual (ctrl); lane NTC: no template control. Note: the exon-19 skipped transcript was detectable only after a semi-nested PCR, performed starting from 1 μL of the original RT-PCR product using a specific primer spanning exons 18–20. Traces of the aberrantly-spliced product were evidenced also in the wild-type sample, indicating that this splicing is present also under physiological conditions. Bottom right: Sequence electropherograms of RT-PCR products confirming the aberrant junction between exons 18/20 (c.2546+3G>C), panel A, and exons 49/51 (c.8155+6T>C), panel B.