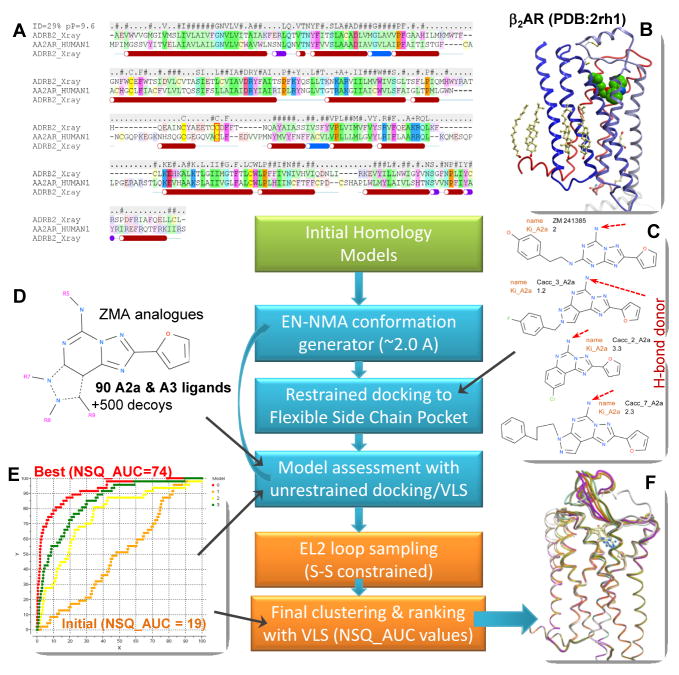
Figure 2.
A flowchart of the modeling algorithm. Initial homology modeling (green block) uses AA2AR/β2AR alignment (A) and β2AR structural template (B). The ligand-guided optimization procedure (cyan blocks) generates multiple conformations of the protein backbone with EN·NMA algorithm, which is followed by docking selected ligands (C) into the models with flexible side chains. Resulting models of receptor are evaluated by fast (rigid) docking of a set of AA2AR ligands and decoys (D), and the models with the best NSQ_AUC values (E) are selected. Final modeling steps (orange blocks) include loop modeling and ranking of the final AA2AR-ZMA models (F).