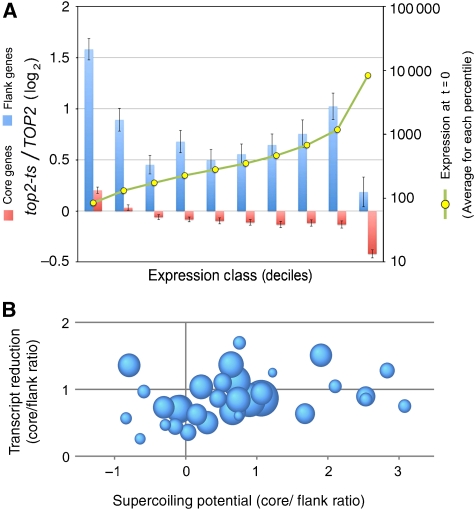
Figure 4.
The positional response to DNA helical stress is not consequent to different levels of transcription activity between chromosomal core and flanking genes. (A) Comparison between the effect of DNA (+) helical stress in chromosome flanking genes (at <50 kb form the telomere) and chromosome core genes (the rest), which are classified by their transcript levels before accumulation of helical stress. The classes are deciles (10 equal groups of about 480 genes) of genes with similar mRNA abundance at time 0 (yellow dots indicate averaged fluorescence units). The resulting graph illustrates that flanking genes (blue) behave always different from core genes (pink), irrespectively of their initial transcript levels. (B) Comparison of the capacity to generate transcription-driven supercoils between chromosomal core and flanking regions. Each of the 32 spheres corresponds to an individual chromosomal arm, being the sphere area proportional to the arm length. The horizontal axis plots the core/flank ratio (log2) of supercoiling potential for each individual arm (see text for details). Note how some flanks can generate more transcription-driven supercoils than their respective core regions. The vertical axis plots the core/flank ratio of transcript level change (log2) for each individual arm after the accumulation of DNA helical stress.