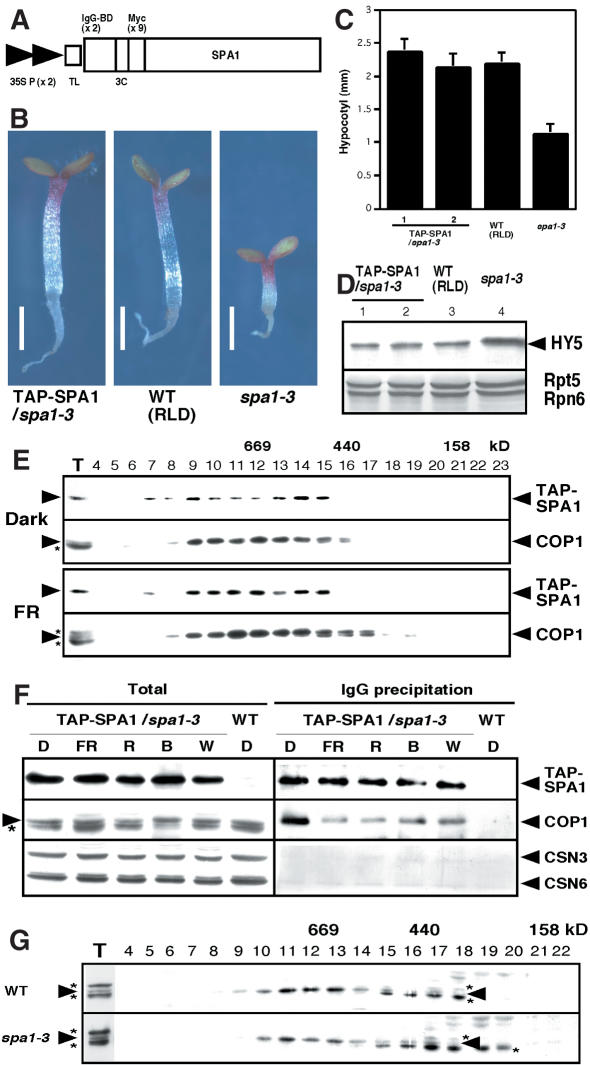
Figure 4.
COP1 interacts with SPA1 in vivo, but light induces their dissociation. (A) The modified TAP-SPA1 construct used for plant transformation, consisting of two copies of the IgG-binding domain (IgG-BD), a human rhinovirus 3C protease cleavage site (3C), and nine copies of the myc epitope tag downstream of two tandem copies of the cauliflower mosaic virus 35S promoter (35S P) with a tobacco mosaic virus translational enhancer sequence (TL). (B-D) TAP-SPA1 complements spa1-3 under continuous far-red light at the level of seedling morphology (B), hypocotyl length (C), and HY5 abundance (D). Bars, 1 mm. Error bars represent the standard deviation of >20 plants each. An immunoblot probed with anti-Rpt5 and anti-Rpn6 antibodies is also shown as a loading control. (E) Gel filtration analyses of 4-day-old dark-grown and continuous far-red-grown TAP-SPA1 transgenic plants. (F) IgG precipitation using the TAP-SPA1 plants grown under various light conditions. (D) Continuous darkness; (FR) continuous far-red light; (R) continuous red light; (B) continuous blue light; (W) continuous white light. (G) Gel filtration profiles of COP1 under continuous far-red light (FRc). The wild-type (WT) ecotype used is RLD. The arrowheads and asterisks in panels D-G indicate protein positions and the anti-COP1 antibody cross-reacting bands, respectively.