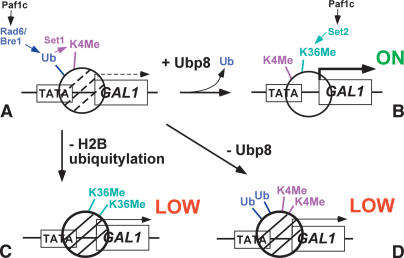
Figure 7.
Model of the role of ubiquitylation and deubiquitylation at GAL1. Schematic representation of the GAL1 promoter and a nucleosome over the TATA/5′ end of the ORF under poised (A; Ub + K4Me) and activated (B; K4Me + K36Me) conditions, showing the relationship between ubiquitylation/deubiquitylation and methylation in the wild-type strain. Paf1 complex loads both Set1 (K4Me) and Set2 (K36Me). The lower cartoons represent altered ubiquitylation and methylation in ubp8Δ and htb1-KR backgrounds. (C) In the absence of H2B ubiquitylation, there is no K4 methylation and high K36 methylation. (D) In the absence of Ubp8, there is high K4 methylation and lowK36 methylation. These altered ubiquitylation/methylation patterns lead to lowtranscription.