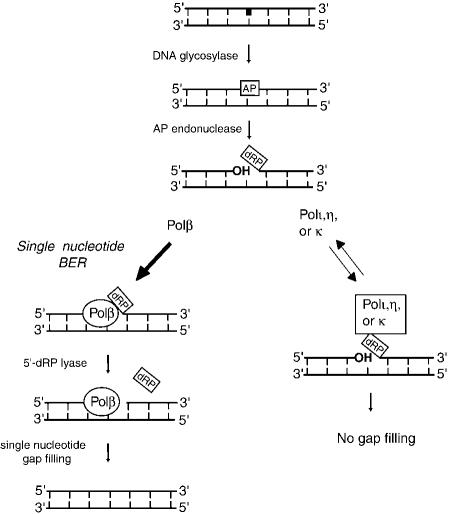
Figure 5.
A model for the exclusion of Y-family polymerases from DNA synthesis during BER. An AP site is generated in DNA either by the action of a DNA glycosylase on the damaged base (shown in bold) or via spontaneous hydrolysis of purines (data not shown). Nicking of the phosphodiester backbone on the 5′-side of the AP lesion by a class II AP endonuclease generates a 3′-OH terminus and a 5′-dRP moiety, which is removed by the potent 5′-dRP lyase activity of Polβ, followed by single nucleotide gap filling by this enzyme (left). The thick arrow on the left denotes that because of its high affinity for the lesion site, Polβ would be the predominant enzyme to carry out the reactions shown. The low-fidelity Y-family polymerases, however, would be excluded from the DNA synthesis reaction during BER because of their unproductive binding to the 5′-dRP residue, which inhibits their ability to synthesize DNA on such a substrate (right). The bidirectional thin arrows on the right signify the less efficient and transient binding of Y-family polymerases to the lesion site. Dissociation of these polymerases from the 5′-dRP residue would free up the residue for removal by the 5′-dRP lyase activity of Polβ and for repair synthesis, as is shown on the left.