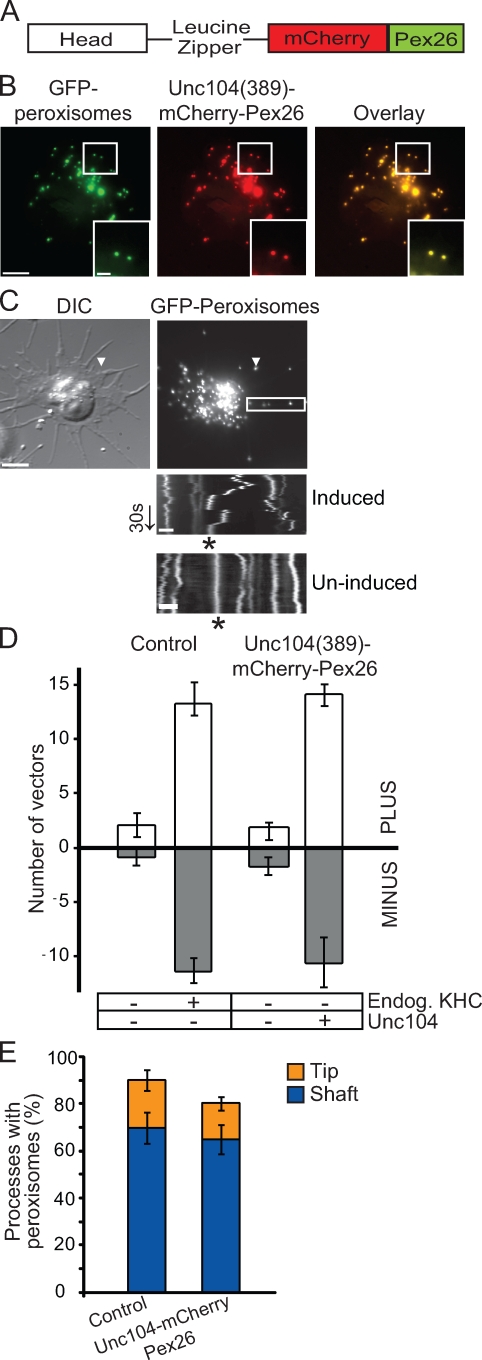
Figure 2.
Unc104 replaces kinesin-1 in bidirectional peroxisome transport. (A) Schematic of Unc104(389)–mCherry-Pex26 construct. Dimeric human Unc104 construct includes the head and the neck-linker domains (amino acids 1–389) followed by a leucine zipper for dimerization. Human Pex26 is fused to mCherry to generate a peroxisome-targeting vector. (B) Representative images showing colocalization of Unc104(389)–mCherry-Pex26 and GFP-labeled peroxisomes in stably transfected S2 cells. Boxed areas are shown at higher magnifications in the insets. (C) Unc104(389)–mCherry-Pex26 restores bidirectional peroxisome motility in KHC-depleted cells. Unc104(389)–mCherry-Pex26 expression induced with 5 mM copper sulfate. Arrowheads highlight location of a single peroxisome within a process. Images corresponds to Video 4. Boxed area delineates the region selected for kymograph analysis (bottom). Top kymograph is from a cell in which Unc104(389)–mCherry-Pex26 expression has been induced. Bottom kymograph is from a cell in which Unc104(389)–mCherry-Pex26 expression has not been induced. Endogenous KHC has been depleted in both cases. Asterisks indicate the track of a peroxisome. DIC, differential interference contrast. Arrow delineates time (30 s). (D) Graph showing the number of peroxisome vectors >0.2 µm in S2 cells expressing Unc104(389)–mCherry-Pex26 in an endogenous KHC-depleted background. Data represent mean values ± SD from 30 cells per condition (from three separate dsRNA treatments). PLUS refers to those peroxisomes moving toward the tips of processes, whereas MINUS refers to those moving toward the cell center. (E) Replacement of endogenous KHC with Unc104(389)–mCherry-Pex26 does not affect peroxisome distribution along processes. Graph representing the percentage of processes containing peroxisomes after induction of Unc104(389)–mCherry-Pex26 in a KHC-depleted background. Data are presented as mean values ± SD from 120 cells (from three separate dsRNA treatments). Bars: (B and C [top]) 5 µm; (C [bottom]) 1 µm.