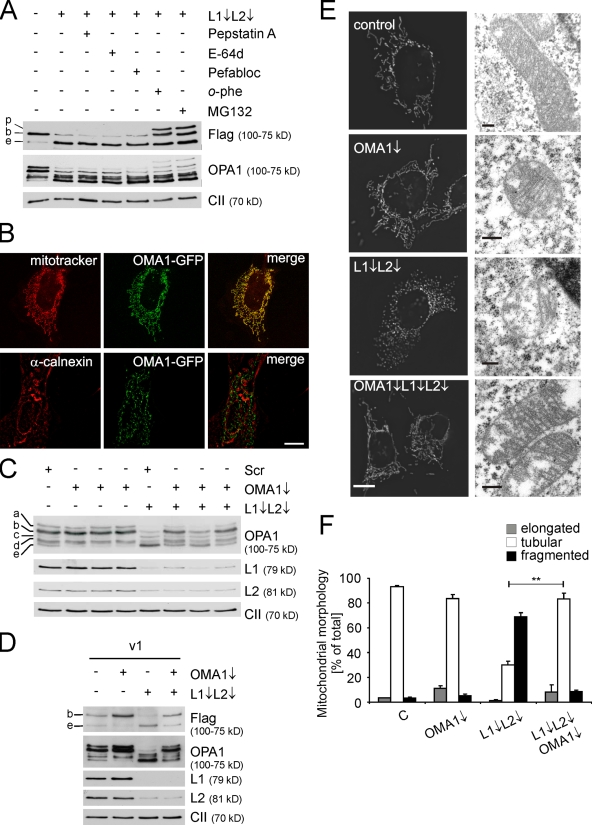
Figure 5.
OMA1 degrades OPA1 in AFG3L1/AFG3L2-deficient MEFs. (A) Stability of OPA1 and OPA1 variant 1 in the presence of different protease inhibitors. Flag-tagged OPA1 variant 1 was expressed in MEFs that were subsequently depleted of AFG3L1 (L1) and AFG3L2 (L2). After 48 h, MEFs were incubated for 5 h with pepstatin A, E-64d, Pefabloc SC, o-phe, or MG132, and cell lysates were analyzed by immunoblotting with α-Flag, α-OPA1, and α–complex II. p, OPA1 precursor; b, long OPA1 isoform; e, short OPA1 isoform. (B) Localization of OMA1 to mitochondria. Fluorescence microscopy of MEFs expressing murine OMA1 harboring a C-terminal GFP tag. Mitochondria were stained with MitoTracker red CMXRos, whereas the ER was detected using an antiserum directed against calnexin. (C) Immunoblot analysis of MEFs transfected with siRNAs directed against AFG3L1 and -2 and OMA1 or scrambled siRNA (Scr) using specific antisera recognizing OPA1, AFG3L1 and -2, and the 70-kD subunit of complex II (CII). Three different siRNAs directed against OMA1 were used. (D) Immunoblot analysis of MEFs transfected with siRNAs directed against AFG3L1/AFG3L2 and OMA1 and Flag-tagged OPA1 splice variant 1 (v1). Cell lysates were analyzed 3 d after transfection by SDS-PAGE and immunoblotting using α-Flag, α-AFG3L1, α-AFG3L2, α-OPA1, and α–complex II antibodies. (E) Mitochondrial morphology in MEFs transfected with AFG3L1/AFG3L2 and OMA1 siRNA was visualized by expression of mito-DsRed (left). Mitochondrial ultrastructure was analyzed by electron microscopy. (F) Quantification of mitochondrial morphology. >150 MEFs were scored in each immunofluorescence experiment. Bars represent means ± SD of three independent experiments (**, P < 0.01). Bars: (B) 10 µm; (E) 0.2 µm.