Fig. 1.
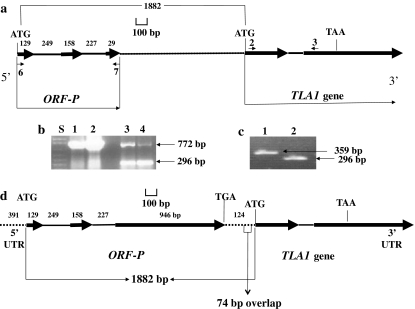
Identification of an incomplete predicted open reading frame (ORF-P) in the Chlamydomonas reinhardtii database. a Genomic map of the ORF-P showing predicted exons (thick black arrow) and introns (thin black lines in-between exons). The uncharacterized genomic DNA region between ORF-P and TLA1 is depicted by a dotted line. The small black arrows (6, 7; 2, 3) denote primers used for genomic DNA PCR and RT-PCR. The known start codon (ATG) and the stop codon (TAA) of the TLA1 gene are denoted. Numbers refer to the base pairs of the putative exons, introns and distance between the start codon of ORF-P and start codon of TLA1. b Lanes 1 and 2 show the genomic DNA PCR product (772 bp) obtained with ORF-P-specific primer 6 and primer 7 (Exon-1/Exon-3 of ORF-P; Table 1); lanes 3 and 4 show the RT-PCR product (296 bp) obtained with the same primer set. (Genomic DNA contamination in the RNA preparation gave rise to the 772 bp product in the RT-PCR reaction in lanes 3 and 4.). Lane S refers to 1 kb plus DNA ladder. c RT-PCR using TLA1 gene-specific Exon-1 “primer 2” and Exon-2 “reverse primer 3R” (359 bp product; lane 1, Table 1) and ORF-P gene-specific Exon-1 “primer 6” and Exon-3 “primer 7” (296 bp product; lane 2, Table 1). d Complete genomic DNA map of the C. reinhardtii ORF-P and TLA1 genes. The positional alignment of the TLA1 gene with respect to the ORF-P is shown. Thick black arrows denote exons, while thin black lines denote introns. Untranslated regions of the two genes are denoted by dotted lines. Note that the 3′ UTR of ORF-P overlaps the 5′ UTR of TLA1 by 74 bp. The start and stop codons of the ORF-P and TLA1 genes are labeled. Numbers denote the base pairs of the respective DNA regions, i.e., 5′ UTR, exons, introns and 3′ UTR of the ORF-P