Figure 3. Conservation of the glutamate switch in AAA+ proteins.
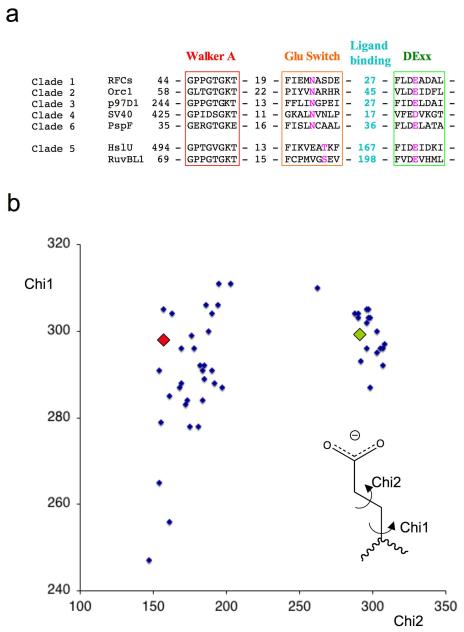
(a) Sequence alignments of selected members of each clade for which crystal structures are known. The colour scheme follows that used in Figure 4. RFCs – Replication factor C small subunit A.fulgidus, Orc1 – Orc1 protein A.pernix, p97D1 - p97 D1 AAA+ domain M.musculus, SV40 – SV40 Large T antigen, PspF – PspF E.coli, HslU – HslU E.coli, RuvBL1 – RuvB-like 1 (TIP49a, Pontin) H.sapiens.
(b) Plot of side chain torsion angles for 50 active site glutamate (DExx box) residues from AAA+ protein structures in the protein database (www.rcsb.org) with a resolution better than 3.5Å. Only one copy from the asymmetric unit was used if angles were similar to reduce redundancy. The appropriate glutamate residue was selected from individual PDB files and the side chain torsion angles were calculated using the CCP4 program ANGLES 40 then normalized to 0-360 degrees for display. The values for the PspF-ADP complex (green diamond) and PspF-ATP complex (red diamond) are overlaid as examples of residues in the two conformations.