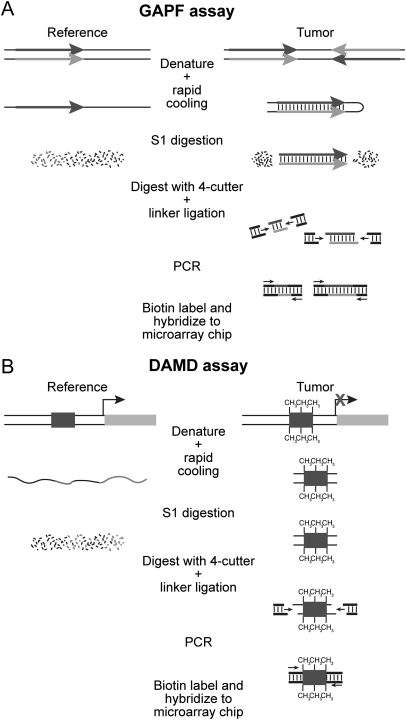
Fig. 1.
Schematic of the genomewide analysis of palindrome formation (GAPF) and denaturation analysis of methylation differences (DAMD) assays. (A) GAPF assay. Palindromic sequences can rapidly anneal intramolecularly to form “snap-back” DNA under conditions that do not favor intermolecular annealing. This snap-back property is used to enrich for palindromic sequences in total genomic DNA by denaturing the DNA at 100°C, rapidly renaturing it in the presence of 100 mM NaCl, and then digesting the mixture with the single-strand-specific nuclease S1. Snap-back DNA formed from palindromes is double stranded and resistant to S1, whereas the remainder of genomic DNA is single stranded and thus is sensitive to S1 digestion. Ligation-mediated PCR is performed, and then the DNA is labeled and hybridized to a microarray for analysis. (B) DAMD assay. DNA manipulations are the same as those performed in GAPF. During the denaturation step of the assay, the increase in the Tm caused by 5-methylcytosine renders methylated DNA resistant to denaturation compared to unmethylated DNA. After rapid cooling, the methylated DNA remains double stranded and resistant to S1 nuclease, whereas the same unmethylated sequence in the reference sample is single stranded and thus sensitive to S1 digestion.