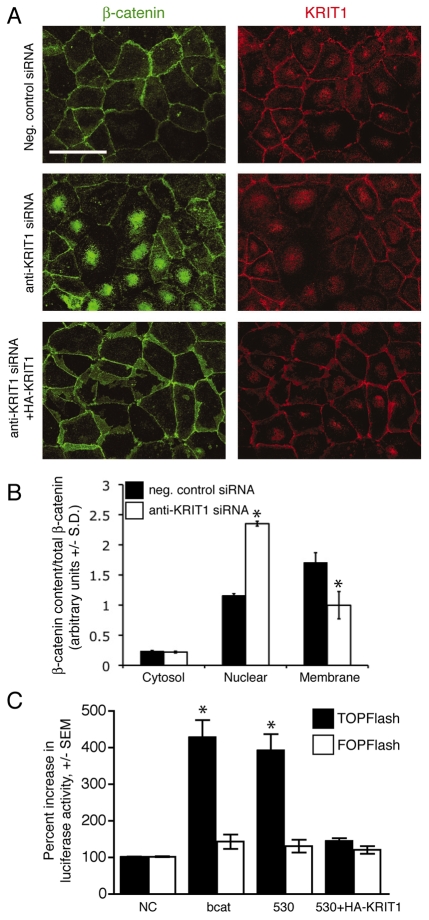
Fig. 2.
Loss of KRIT1 regulates β-catenin nuclear localization and activatesβ-catenin-dependent transcription. (A) β-Catenin and KRIT1 staining of confluent BAECs. Top panels, negative control siRNA; center panels, anti-KRIT1 siRNA; lower panels, anti-KRIT1 siRNA co-transfected with siRNA-resistant KRIT1. Confocal images are representative, n=6. Bar, 50 μm. (B) Densitometric quantification of subcellular fractionation. The data shown are the representative expression of β-catenin in cytoplasmic, nuclear and membrane fractions of siRNA-transfected HPAECs, and are relative to total β-catenin ±S.D. *P<0.05 by Student’s two-tailed t-test, n=3. Representative blots for β-catenin and KRIT1 expression, as well as fractionation control blots, can be found in supplementary material Fig. S1A. (C) TOPFlash reporter activity in KRIT1-depleted cells. Data are shown relative to negative control siRNA-transfected cells (NC). β-cat, FLAG–β-catenin expressing cells; 530, anti-KRIT1 siRNA expressing cells; 530+HA-KRIT1, anti-KRIT1 siRNA and hemagglutinin-tagged (HA)-KRIT1 expressing cells. Overall P=0.002 by analysis of variance (ANOVA), *P<0.05 by Bonferroni post-hoc comparison to negative control siRNA-transfected cells, n=5.