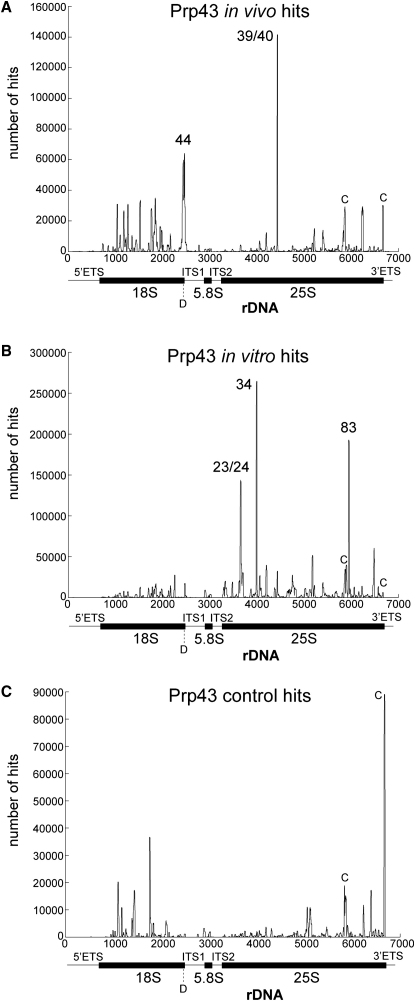
Figure 1.
Identification of Putative Binding Sites of Prp43 on Ribosomal RNA
Control cells (C) or Prp43-HTP-expressing cells (A) were UV crosslinked in vivo or crosslinking was performed in vitro after cell lysis and enrichment of Prp43-HTP-containing complexes on IgG Sepharose (B). Crosslinked RNAs were trimmed and ligated to linkers followed by RT-PCR and Solexa sequencing. The obtained sequences were aligned with the rDNA encoding 35S pre-rRNA (nucleotides 1–6880), and the number of hits for each individual nucleotide is plotted. The position of the mature 18S, 5.8S, and 25S rRNAs is indicated by bars below, and processing site D is shown. Numbers at peaks indicate the corresponding helices of 18S or 25S, while 25S peaks labeled with “C” represent background peaks also found in the control. In vivo crosslinking of Prp43-HTP-expressing cells resulted in two major peaks, whereas in vitro crosslinking of purified complexes yielded three sequence clusters. Thus, CRAC experiments have identified several potential binding sites for Prp43 on pre-rRNA.