Figure 2.
Interactions with rRNA and mRNA
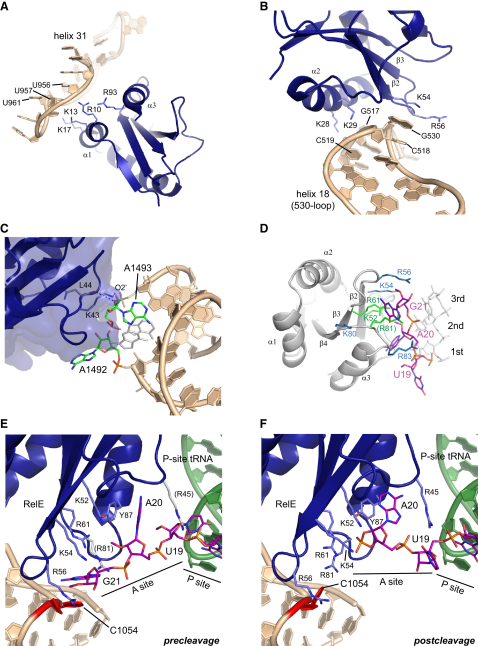
(A) Interactions between RelE (blue) and the head region of the 30S (helix 31, wheat). (A)–(E) are based on the precleavage structure. All rRNA references correspond to the E. coli sequences.
(B) Interactions between RelE and the body region of the 30S (helix 18, wheat).
(C) Interactions between RelE (blue, semitransparent surface with interacting residues shown as sticks) and the decoding center bases A1492/A1493 (green sticks) of 16S rRNA helix 44 (wheat). The unbound conformation of the decoding site is shown with white sticks.
(D) Overview of the contacts between RelE and mRNA. RelE is shown as a white cartoon with basic residues near the mRNA as blue sticks. Residues known to affect the activity of P. horikoshii RelE are shown in green. The normal mRNA path is shown with white sticks and the RelE-bound mRNA path with purple sticks. The position of the R81 side chain is inferred from the Cβ position.
(E and F) Details of the interactions with mRNA in the precleavage (E) or post-cleavage (F) state showing RelE (blue) with relevant side chains as sticks, P site tRNA (green cartoon), the 16S conserved rRNA helix 34 bulge (wheat), and the mRNA (purple sticks) with the A and P site codons labeled. C1054 is shown in red. The positions of the side chains of R45 and R81 are inferred from Cβ positions and are therefore shown in gray.
See also Figure S2.