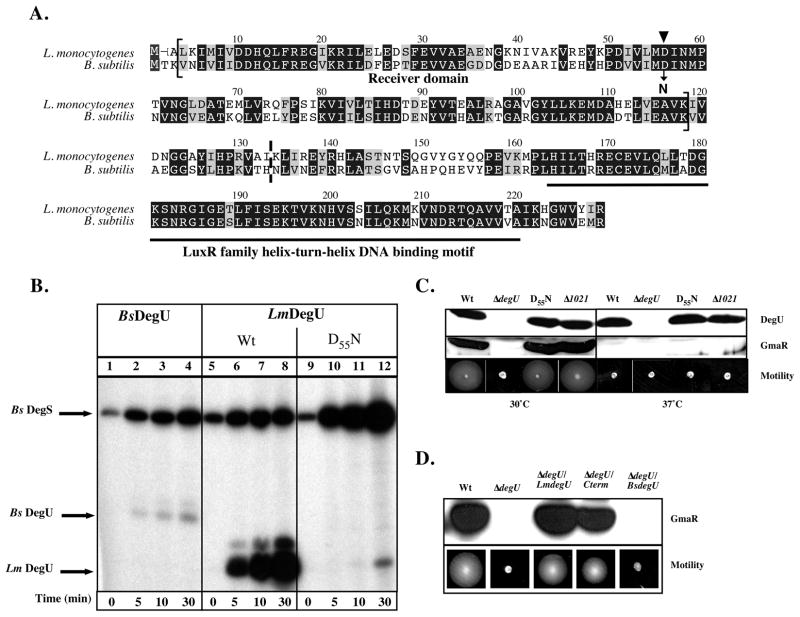
Figure 1. Phosphorylation of DegU.
A. Alignment of amino acid residues of LmDegU and BsDegU proteins. Conserved identical residues are blocked in black and similar residues are blocked in grey. The LmDegU phosphorylation receiver domain (aa 3-117) is indicated by brackets and contains the conserved phosphoryl acceptor site D55 (▼). The LuxR/FixJ family helix-turn-helix motif (aa 162-219) is underlined in black. The N-terminal truncation site in ΔdegU/Cterm is marked by a dashed line between aa 132 and aa 133.
B. in vitro phosphorylation of DegU. Purified BsDegS sensor kinase was incubated with either BsDegU (lanes 1–4), wild-type (Wt) LmDegU (lanes 5–8), or LmDegUD55N (lanes 9–12) in the presence of [γ32P]-ATP. LmDegUD55N has a phosphoryl acceptor site mutation Asp55 to Asn. Reactions were incubated at room temperature (RT) 5, 10, or 30 min. Proteins were resolved by SDS-PAGE and phosphorylation was detected using autoradiography.
C. Western blot and motility analysis of DegUD55N and Δlmo1021 bacteria. DegU and GmaR protein levels were examined using Western blot analysis of whole cell lysates prepared from cultures grown at either 30°C or 37°C for 18 h. A GmaR- or DegU- specific polyclonal antibody was used for detection. Motility was examined in low agar (0.3%) motility plates inoculated with a straight needle and incubated for 48 h.
D. Western blot and motility analysis of ΔdegU/Cterm and ΔdegU/BsDegU bacteria. A DegU-negative strain was complemented with either full-length LmDegU (ΔdegU/LmdegU), a N-terminally truncated DegU (ΔdegU/Cterm), or full-length BsDegU (ΔdegU/BsdegU). Whole cell lysates were prepared from cultures grown at 30°C for 18 h. Western blot analysis was used to detect GmaR using a GmaR-specific polyclonal antibody. Motility was also examined at 30°C in low agar (0.3%) motility plates inoculated with a straight needle and incubated for 48 h.