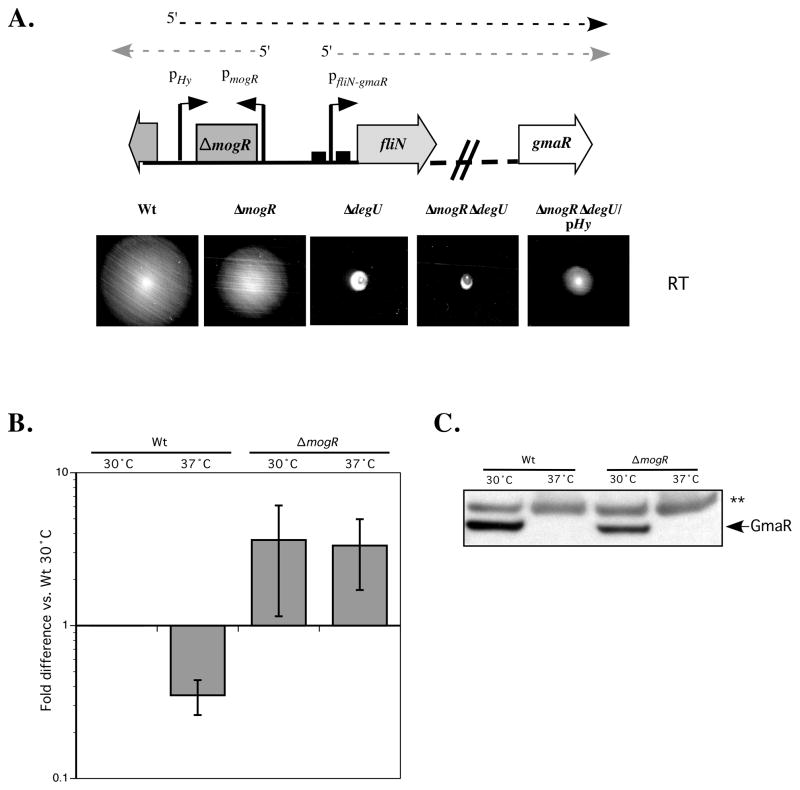
Figure 5. A post-transcriptional mechanism controls temperature-dependent production of GmaR.
A. Constitutive expression of the fliN-gmaR operon in ΔmogRΔdegU. Native promoters for mogR and fliN-gmaR are represented by bent arrows (pmogR and pfliN-gmaR). MogR binding sites are marked as boxes overlapping the fliN-gmaR promoter. Divergent transcripts initiating from the native promoters are drawn as light dashed arrows. The pHy promoter was inserted upstream of the native fliN-gmaR promoter in a ΔmogRΔdegU strain to constitutively drive expression of the fliN-gmaR operon. Transcription initiating from this promoter is drawn as a dark dashed line. For the motility assay analysis, strains were inoculated into low agar (0.3%) motility plates with a straight needle and incubated at RT for 48 h.
B. Real-Time quantitative PCR analysis of gmaR transcripts. RNA was extracted from wild-type (Wt) and ΔmogR strains grown at either 30°C or 37°C for 24 h. Samples were DNaseI treated and reverse-transcribed with random hexamers to generate cDNA. Relative gene expression was quantified by using Real-Time PCR and the Pfaffl method (2−ΔΔCT). Results represent the average and standard deviation of three independent experiments. The iap gene was used as an internal standard and the Wt 30°C sample was set as the calibrator.
C. GmaR protein analysis by Western blot. Total protein samples from cultures used in B were processed for SDS-PAGE and Western blot analysis. A GmaR-specific polyclonal antibody was used for detection. ** indicates a non-specific band that is shown as a loading control.