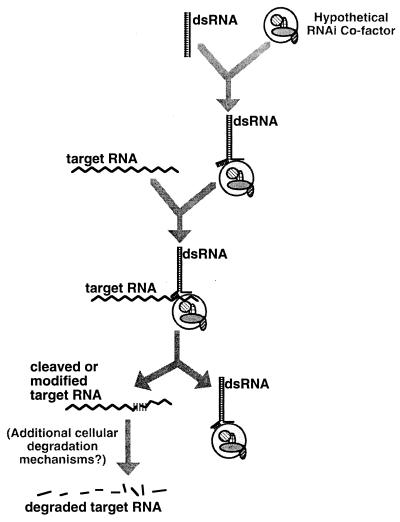
Figure 4.
Possible model for dsRNA-mediated genetic interference in C. elegans. Upon introduction into the cell, dsRNA is proposed to complex with a (hypothetical) protein or ribonucleoprotein complex that allows unwinding of an arbitrary segment of the duplex. The complex would then search by homology for corresponding segments of cellular RNA. Recognition of cellular RNAs would be followed by a process marking the target RNA for degradation. Possible “marking” mechanisms include direct cleavage of the target RNA, covalent modification (e.g., by adenosine deaminase), or the recruitment or removal of specific RNA-binding proteins. Whatever the mechanism of this initial interaction, our data suggest a rapid subsequent degradation of the entire targeted transcript. The secondary degradation machinery could involve a combination of components specific to RNA interference and/or more general catabolic mechanisms.