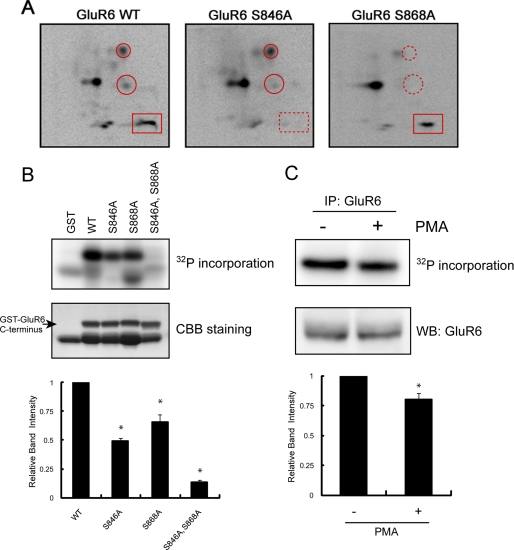
FIGURE 1.
Identification of Ser846 and Ser868 as PKC phosphorylation sites in the GluR6 C terminus. A, phosphopeptide map analysis shows that the GluR6 C terminus is phosphorylated by PKC on multiple peptides (left panel) including Ser846 and Ser868, as indicated by the disappearance of phosphopeptides on maps of the GluR6 mutants (middle and right panels). GST-GluR6 (WT, S846A, and S868A) fusion proteins were phosphorylated with purified PKC in vitro using [γ-32P]ATP and then subjected to two-dimensional phosphopeptide mapping as described under “Experimental Procedures.” A rectangle surrounds the phosphopeptide that includes phosphorylated Ser846. Circles surround the phosphopeptides that include phosphorylated Ser868. An X in each panel indicates the origin where the peptides were initially spotted. Representative phosphopeptide maps are shown of 3–4 independent experiments. B, Ser846 and Ser868 are the major PKC phosphorylation sites in the GluR6 C terminus. GST, GST-GluR6 WT, GST-GluR6 S846A, GST-GluR6 S868A, and GST-GluR6 S846A,S868A were phosphorylated by PKC in vitro and analyzed by PhosphorImager. The loaded amount of GST fusion protein was visualized by protein staining. The lower band in the WT lane is a degradation product of GST-GluR6. Data represent mean ± S.E.M. (error bars) of normalized relative band intensity. *, p < 0.05, Student's t test. CCB, Coomassie Brilliant Blue. C, pretreatment of neurons with PMA reduced PKC phosphorylation of GluR6 in vitro. GluR6 was immunoprecipitated from PMA or vehicle-treated neurons, phosphorylated by PKC in vitro, and analyzed by PhosphorImager. The loaded amount of GluR6 was visualized by Western blotting using anti-GluR6 antibody. Data represent mean ± S.E.M. of normalized relative band intensity. *, p < 0.05, Student's t test.