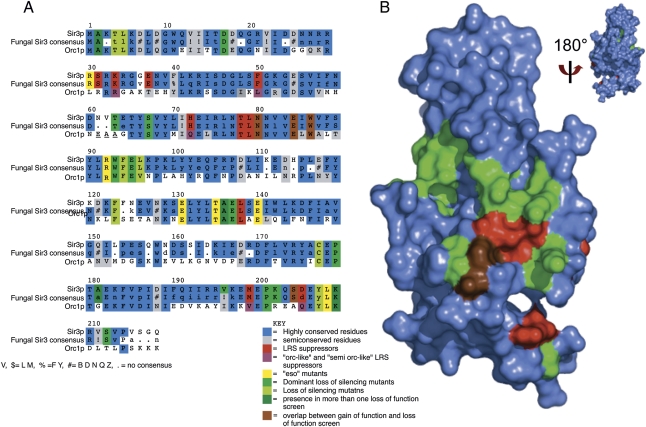
Figure 2.
Critical binding surface of the BAH domain revealed by multiple genetic screens. (A) Multiple sequence alignment of SIR3 BAH domain from S. cerevisiae, consensus sequence for SIR3 BAH domain from post-genome duplication yeast strains, and BAH domain from ORC1. The side chains identified in the eso, DNG, and lfm screens are highlighted in yellow-green, medium green, and light green, respectively; side chains from the slr screen are highlighted in red; residues overlapping between the slr screens and other loss-of-function screens are highlighted in brown; residues overlapping among the loss-of-function screens DNG, eso, and lfm are highlighted in dark green. Mutations in the slr screen that introduce an “Orc-like” residue are highlighted in purple. Conserved residues are highlighted in blue, and semiconserved residues are highlighted in gray. (B) Surface representation of the crystal structure 2FL7 (Hou et al. 2006). The color coding is similar to A, but simplified; the DNG, eso, and lfm screens are highlighted in green; the slr screen is highlighted in red; and the overlap between the two groups is highlighted in brown. The structure is rotated 180° around the X-axis to demonstrate that all of the side chains identified by the various screens are found on one face of the BAH domain.