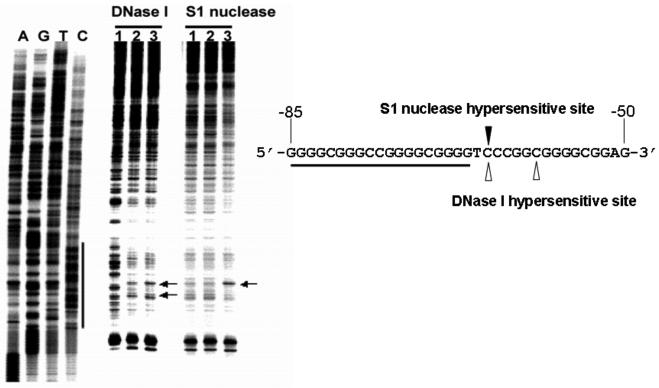
Fig. 3.
In vitro footprinting of the VEGF promoter region with DNase I, S1 nuclease or DMS. Autoradiograms showing S1 nuclease and DNase I cleavage sites on the G-strand of a supercoiled pGL3-V789 plasmid. This plasmid was incubated in control (lane 1), or in KCl buffer without (lane 2) and with 1 μM telomestatin (lane 3) at 37°C for 1 h before digesting with nucleases. Arrows indicate the hypersensitive cleavage sites to nucleases. The primer extension reaction revealed a long protected region at approximately −53 to −123 bp, including the G-rich sequences, when a supercoiled pGL3-V789 plasmid was incubated with 100 mM KCl and digested with DNase I (compare lanes 1 and 2). This indicates a possible transition from B-DNA to a G-quadruplex structure in the VEGF promoter region, which is consequently resistant to DNase I digestion. Significantly, a striking hypersensitivity was found in the presence of KCl and telomestatin at a cytosine located at the 3′-side of the G-quadruplex-forming region (underlined sequence), which is the junction site separating the putative G-quadruplex from the adjacent normal B-DNA (see arrow “A” in lane 3). The reactivity of S1 nuclease at the VEGF proximal promoter region was also moderately reduced in the presence of telomestatin and KCl, and the hypersensitivity site observed with S1 nuclease corresponds to one of those obtained with DNase I in the presence of telomestatin and KCl (lane 3) (Figure modified from ref. 30).