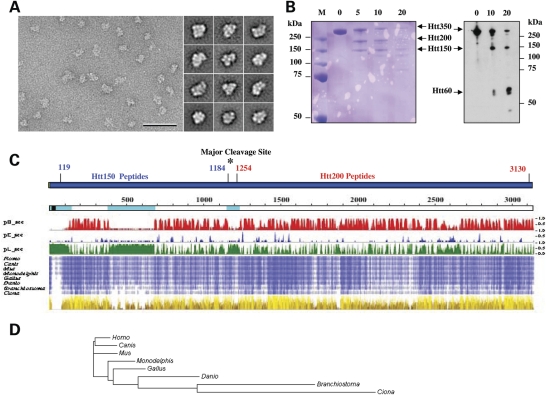
Figure 1.
Huntingtin conformational flexibility and domain organization. (A) Representative area of an electron micrograph of negatively stained FLAG-Q23 huntingtin (scale bar 50 nm) (left) and representative class averages (side length of panels is 28.8 nm) (right) showing the structural variability of huntingtin. (B) Timed proteolysis of Q23 FLAG-huntingtin (Htt350) with trypsin (0, 5, 10, 20 min) yielded two major Coomassie Blue stained products at ∼150 kDa (Htt150) and ∼200 kDa (Htt200) (left), though immunoblot probed with anti-FLAG detected the ∼150 kDa fragment and a smaller ∼60 kDa product (right). (C) Schematic of human huntingtin (blue bar), with FLAG-tag (green) and locations of Htt150 and Htt250 mass spectrometry peptides (Supplementary Material, Tables S1 and S2) and major trypsin cleavage site (asterisk). Below this, huntingtin (open line) is depicted with the polyglutamine tract (black block) and amino acid coordinates (above), NORSp predicted disordered regions (light blue), matching the predictions of PROF where pHsec, pEsec and pLsec represent the probability (1 = high, 0 = low) for helix (red), strand (blue) and neither helix nor strand (green). Below this, compressed multiple sequence alignment of huntingtin from human and seven chordates with increasing intensity of blue shading for residues identical in 4 to 8 organisms and physico-chemical properties (from Jalview) conserved for each amino acid position, from dark brown (least) to bright yellow (most), with height corresponding to increasing conservation. (D) Phylogram based upon alignment of huntingtin homologues for the eight representative chordates, with branch lengths proportional to the inferred evolutionary change.