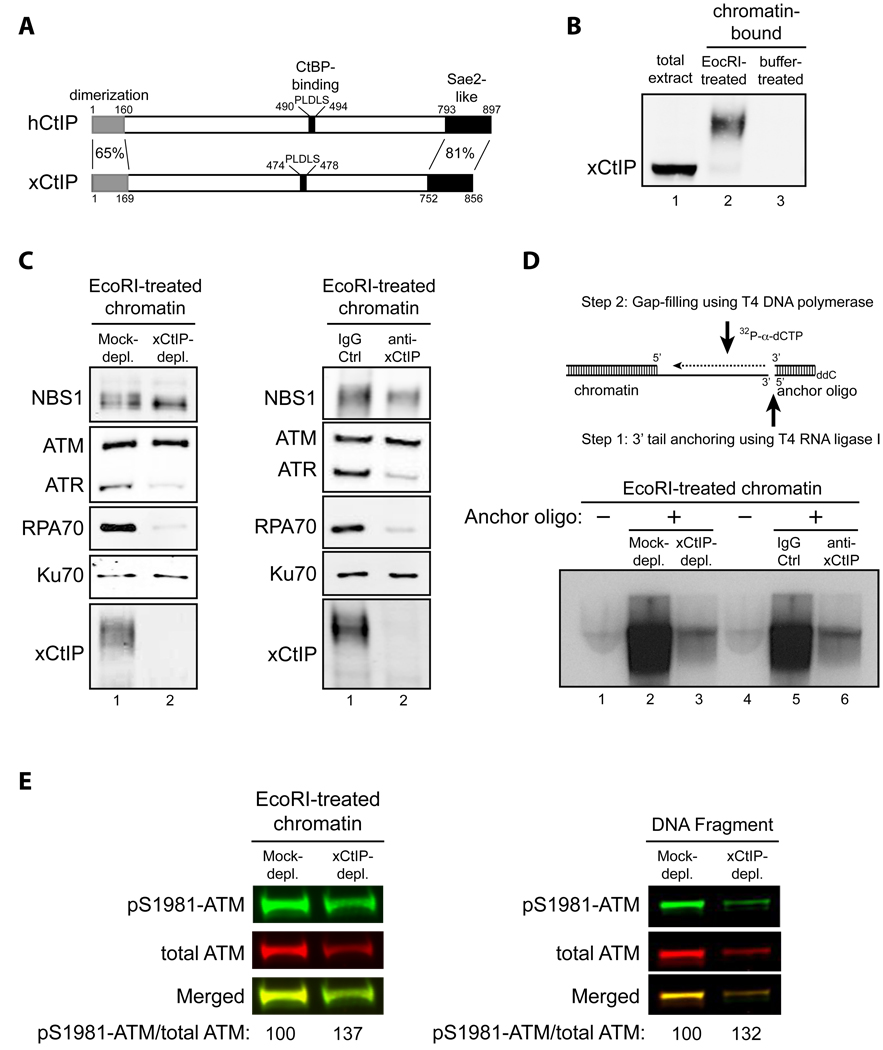
Fig. 1. CtIP is required for DNA end resection, but not ATM activation, in response to DSBs in Xenopus extracts.
- Schematic comparison of human CtIP and Xenopus CtIP (xCtIP).
- xCtIP associated with damaged, but not undamaged, chromatin in the Xenopus extract, and chromatin-associated xCtIP was highly modified as evidenced by its reduced gel mobility. Xenopus extract (20 µl) was incubated with 10,000 EcoRI-treated or buffer-treated demembranated sperm nuclei/µl for 30 min. After incubation, chromatin was isolated and chromatin-associated xCtIP was detected by immunoblotting (lanes 2 and 3). Lane 1, xCtIP in 1 µl of total extract.
- xCtIP is required for association of RPA and ATR, but not NBS1, ATM and Ku70, with chromatin containing DSBs. Left panel: EcoRI-treated sperm chromatin was incubated with mock-depleted or xCtIP-depleted extract for 30 min. After incubation, chromatin was isolated and chromatin associated proteins were detected by immunoblotting. xCtIP depletion blocked chromatin association of RPA70 and ATR, but not NBS1, ATM and Ku70. Right panel: Extract supplemented with 400 ng/µl of control IgG or affinity-purified, neutralizing xCtIP antibodies was incubated with EcoRI-treated sperm chromatin for 30 min. After incubation, chromatin was isolated and chromatin associated proteins were detected by immunoblotting. Addition of xCtIP antibodies inhibited chromatin association of xCtIP, RPA and ATR, but not NBS1, ATM and Ku70.
- xCtIP is required for efficient DSB end resection in the Xenopus extract. Upper panel: A procedure for detecting DSB end resection in the Xenopus extract. The 3’ tail generated by 5’ strand resection of a DNA DSB in chromatin is ligated to the 5’ phosphorylated end of an anchor oligo using T4 RNA ligase I. After ligation, a second, complementary oligo is annealed to the anchor oligo and T4 DNA polymerase is used to fill the gap between the 3’ end of the second oligo and the 5’ resected end in chromatin in the presence of 32P-α-dCTP (see Experimental Procedures in Supplemental Information for details). The amount of 32P-α-dCTP incorporation represents the extent of DSB resection. Lower panels: EcoRI-treated sperm chromatin was incubated with mock-depleted (lane 2) or xCtIP-depleted extract (lane 3), or with extract supplemented with 400 ng/µl of control IgG (lane 5) or neutralizing xCtIP antibodies (lane 6) for 30 min. After incubation, chromatin was isolated and DSB resection was analyzed on agarose gels after oligo anchoring and gap filling in the presence of 32P-α-dCTP. The anchor oligo was omitted from the control lanes 1 and 4.
- Immunodepletion of xCtIP from the Xenopus extract increased the extent of ATM autophosphorylation at S1981 induced by EcoRI-treated sperm chromatin or by dsDNA fragments, although xCtIP depletion partially removed ATM from the extract. Mock-depleted or xCtIP-depleted extract was incubated with 10,000 sperm nuclei/µl (left panel) or 5 ng/µl of a 2 kbp dsDNA fragment (right panel) for 25 min. After incubation, immunoblotting was performed to detect activated ATM and total ATM in the extracts with rabbit phospho-S1981 specific anti-ATM antibodies and guinea pig anti-Xenopus ATM antibodies, respectively. Primary antibodies were detected by Alexa Fluor 680-labeled anti-rabbit secondary antibodies and IRdye 800-labeled anti-guinea pig second antibodies using Odyssey infrared imaging system. Immunoblotting signals were quantified using Odyssey 3.0 software.