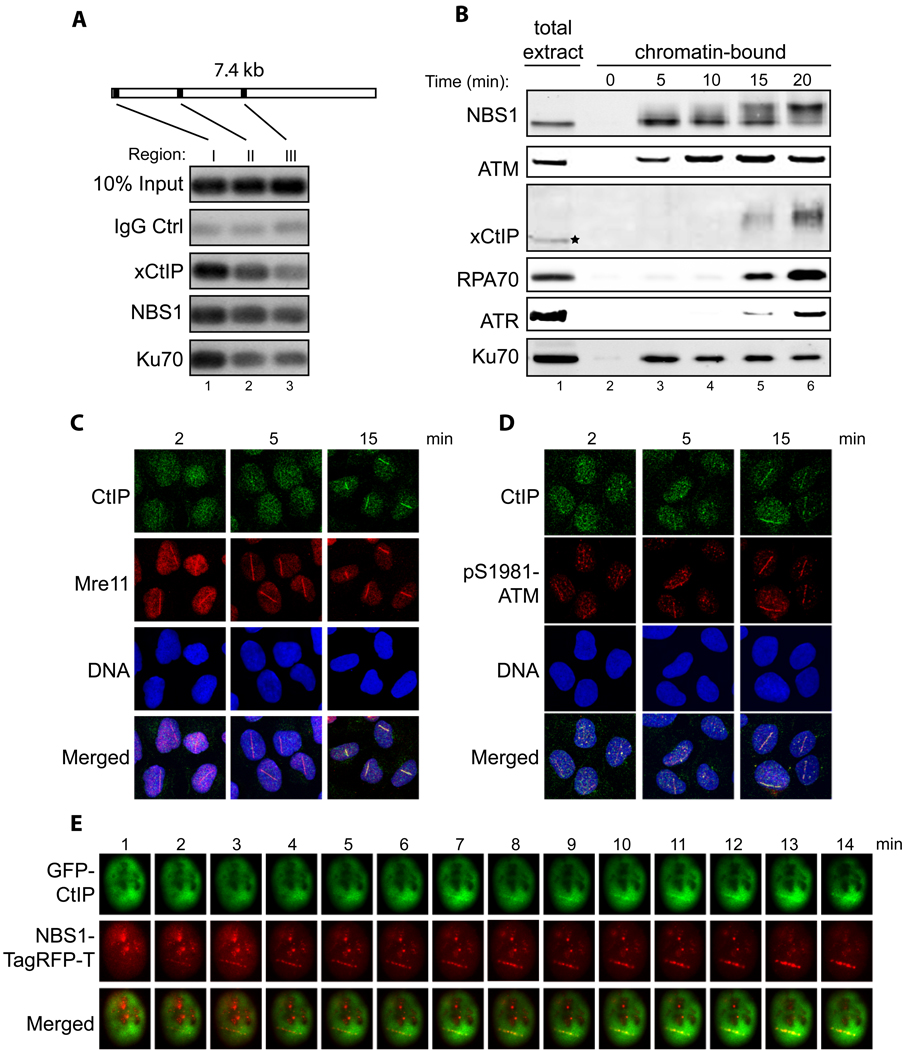
Association of xCtIP, NBS1 and Ku70 with different regions of a DNA fragment incubated in the Xenopus extract. A 7.4 kbp linearized plasmid (5 ng/µl) was incubated with the extract for 15 min. After incubation, ChIP experiments were performed to assess the association of proteins at three representative regions of the DNA fragment: 97–298 bp (region I), 1857–2060 (region II) and 3711–3920 (region III).
Time-course analysis of chromatin association of NBS1, ATM, xCtIP, RPA70, ATR and Ku70 in the Xenopus extract in response to DSBs. EcoRI-treated chromatin incubated in the Xenopus extract was isolated at the indicated times and associated proteins were detected by immunoblotting. Signal in lane 1 represents NBS1, ATM, RPA70, ATR or Ku70 in 0.5 µl of total extract, or xCtIP in 0.1 µl of total extract.
Accumulation of CtIP and Mre11 at DNA damage sites in human cells at different times after DNA damage. Laser irradiation was performed to generate DSBs in a line pattern in human U2OS cells. Subsequently, cells were fixed at the indicated times followed by indirect immunofluorescence to assess the recruitment of CtIP and Mre11 to DNA damage sites. DNA was stained with DAPI.
Same as depicted in panel C, except that the accumulation of CtIP and pS1981-ATM at DNA damage sites was assessed at the indicated time points after laser irradiation.
Laser irradiation was performed in a U2OS cell co-transfected with GFP-CtIP and NBS1-TagRFP-T followed by live cell imaging to assess the kinetics of damage recruitment of CtIP and NBS1 to a damage line.