Figure 2.
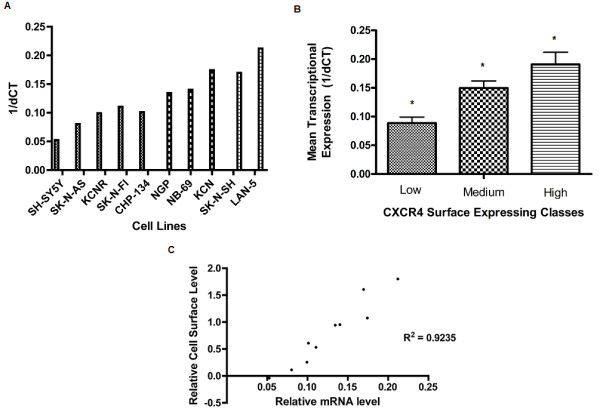
Transcriptional Expression of CXCR4 in Neuroblastoma Cells. Total RNA was isolated from each cell line and used to generate cDNA templates. Quantitative PCR was performed using a total of 250 ng of template cDNA for each cell line and 30 cycles of amplification. A) CXCR4 mRNA levels in panel of ten neuroblastoma cell lines. dCt values for CXCR4 were normalized to the endogenous control glyceraldehyde phosphate dehydrogenase. dCt values represent the average of three replicates. B) The mean dCt value for each surface expression class was generated from all dCt values of cell lines in a given class. Result shows the mean dCt +/- SD of each class to be significantly different from one another (P < 0.005, One-way ANOVA). C) The log10 of mean fluorescent intensity was plotted against the mean dCt for each cell line. Each data point represents the mean value obtained from three FACS assays plotted against the mean value obtained from three quantitative PCR assays. Figure shows a strong and significant correlation between the two forms of expression (Pearson correlation, R2 = 0.9235, P < 0.05).
