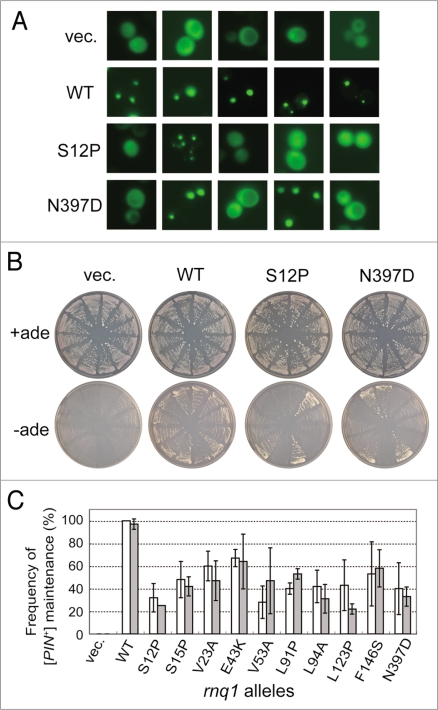
Figure 4.
Quantitative analysis of defects of rnq1 mutants in [PIN+] propagation. (A) Fluorescence microscopy was performed to determine the [PIN+] or [pin−] status of each colony using a Rnq1-GFP reporter. Upon substitution of mutant Rnq1s for the wild-type Rnq1 by plasmid shuffling in strain NS43 ([psi−] [PIN+]), three sets of 20 independent colonies were randomly chosen and again transformed with pRS413CUP1p-Rnq1-GFP to visualize the [PIN+] or [pin−] state with Rnq1-GFP fusion protein. Rnq1-GFP expression under the control of the CUP1 promoter was induced by 50 µM CuSO4 for 3 days on SC plates. Panels show five randomly chosen fluorescent images of each Rnq1 sample. Samples: vec., an empty plasmid; WT, wild-type Rnq1; S12P and N397D, mutant Rnq1s. (B) Pin+ activity monitored by de novo appearance of [PSI+] colonies. Three sets of 12 independent plasmid-shuffled colonies were transformed with plasmid pRS413CUP1p-SUP35NM, and upon overexpression of the Sup35NM domain, these cells were grown on SC+ade (control) and SC-ade plates for seven days. (C) The frequency of appearance of Rnq1-GFP foci (open boxes) and de novo induction of [PSI+] (gray boxes) is expressed as the mean of three independent experiments with standard deviations.