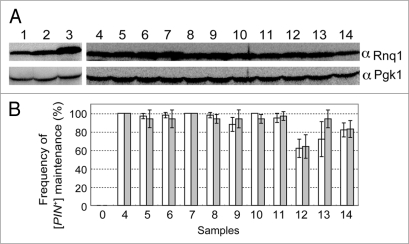
Figure 6.
Suppression of defective rnq1 mutants by overexpression of mutant Rnq1 proteins. (A) Overexpression of Rnq1 proteins examined by western blotting. Experimental conditions and procedures are the same as described in Figure 2 except that rnq1 alleles are expressed from the strong ADH promoter substituted for the authentic RNQ1 promoter. Yeast strains used were NS43 ([psi−] [PIN+]) derivatives whose chromosomal RNQ1 allele is either wild type (lane 1) or nullified with rnq1::KanMX (lanes 2 through 14). Plasmids: 1, plasmid free control; 2, pRS413RNQ1p-RNQ1; 3 and 4, pRS413ADHp-RNQ1; 5, pRS413ADHp-S12P; 6, pRS413ADHp-S15P; 7, pRS413ADHp-V23A; 8, pRS413ADHp-E43K; 9, pRS413ADHp-V53A; 10, pRS413ADHp-L91P; 11, pRS413ADHp-L94A, 12, pRS413ADHp-L123P; 13, pRS413ADHp-F146S; 14, pRS413ADHp-N397D. (B) Frequency of appearance of Rnq1-GFP foci (open boxes) and de novo induction of [PSI+] (gray boxes). Experimental conditions and procedures are the same as described in Figure 5. Three sets of 20 independent colonies were chosen for observation of Rnq1-GFP fluorescence and 12 for monitoring Pin+ activity generated by plasmid shuffling for each rnq1 allele were examined and scored. Results are expressed as the mean of three independent experiments with standard deviations.