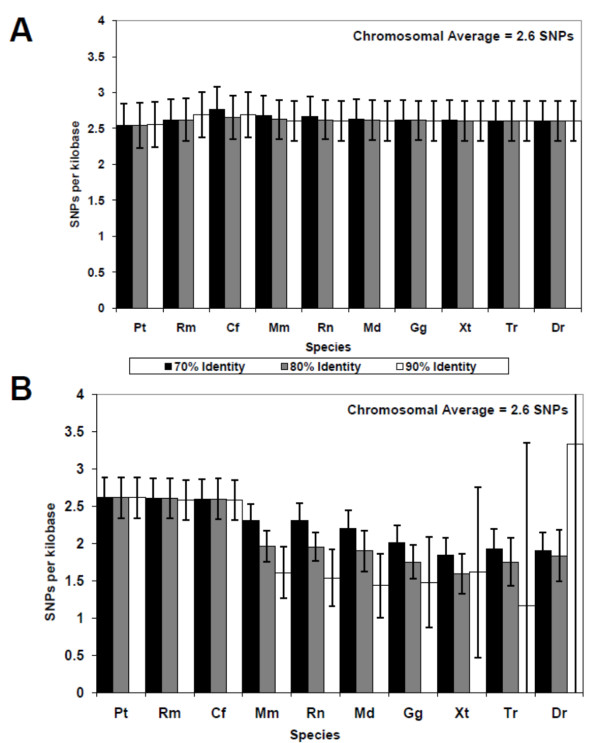
Figure 2.
Bar charts demonstrating SNP densities with the conserved (A) and non-conserved (B) regions of the human genome as determined by pairwise alignment with nine other species (Pt; Pan troglodytes, Rm; Rhesus Macaque, Cn; Canis familiaris, Mm; Mus musculus, Rn; Rattus novergicus, Md; Monodelphis domestica, Gg; Gallus gallus, Xt Xenopus tropicalis, Dr; Danio rerio) at three different selective stringencies (70%; black bars, 80% grey bars; 90%; white bars over 100 base pairs) showing the number of SNPs per kilobase (y-axis) plotted against species (x-axis) that increases in evolutionary "depth" from left to right.