Table 1.
Number of environments (E
opt) and number of replications per environment (R
opt) maximizing the power of QTL detection 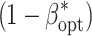 at an experiment-wise error rate of 0.01
at an experiment-wise error rate of 0.01
| Assumptions | Optimum allocation |
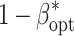 (SE) (SE) |
|||||
|---|---|---|---|---|---|---|---|
| B | l | VC | E opt | R opt | N opt | h 2opt | |
| 1.25 | 25 | 1 | 7 | 3 | 799 | 0.944 | 0.995 (0.0016) |
| 2 | 13 | 3 | 573 | 0.940 | 0.993 (0.0017) | ||
| 3 | 16 | 3 | 486 | 0.906 | 0.982 (0.0026) | ||
| 50 | 1 | 10 | 2 | 752 | 0.952 | 0.996 (0.0009) | |
| 2 | 16 | 2 | 563 | 0.941 | 0.992 (0.0014) | ||
| 3 | 19 | 3 | 411 | 0.919 | 0.970 (0.0026) | ||
| 100 | 1 | 10 | 2 | 752 | 0.952 | 0.996 (0.0006) | |
| 2 | 13 | 3 | 573 | 0.940 | 0.990 (0.0011) | ||
| 3 | 19 | 2 | 484 | 0.905 | 0.938 (0.0035) | ||
| 2.5 | 25 | 1 | 16 | 1 | 1,654 | 0.955 | 0.999 (0.0007) |
| 2 | 16 | 1 | 1,654 | 0.914 | 0.998 (0.0010) | ||
| 3 | 16 | 2 | 1,391 | 0.889 | 0.996 (0.0013) | ||
| 50 | 1 | 10 | 2 | 1,692 | 0.952 | 0.999 (0.0005) | |
| 2 | 10 | 2 | 1,692 | 0.909 | 0.998 (0.0007) | ||
| 3 | 19 | 3 | 1,074 | 0.919 | 0.995 (0.0010) | ||
| 100 | 1 | 7 | 2 | 1,875 | 0.933 | 0.998 (0.0005) | |
| 2 | 16 | 1 | 1,654 | 0.914 | 0.997 (0.0005) | ||
| 3 | 19 | 3 | 1,074 | 0.919 | 0.993 (0.0004) | ||
| 5 | 25 | 1 | 7 | 2 | 3,891 | 0.933 | 1.000 (0.0004) |
| 2 | 16 | 3 | 2,629 | 0.950 | 1.000 (0.0004) | ||
| 3 | 19 | 2 | 2,828 | 0.905 | 0.999 (0.0006) | ||
| 50 | 1 | 7 | 3 | 3,587 | 0.944 | 1.000 (0.0003) | |
| 2 | 10 | 2 | 3,571 | 0.909 | 0.999 (0.0005) | ||
| 3 | 19 | 3 | 2,401 | 0.919 | 0.999 (0.0005) | ||
| 100 | 1 | 10 | 3 | 3,209 | 0.960 | 1.000 (0.0001) | |
| 2 | 10 | 2 | 3,571 | 0.909 | 0.999 (0.0004) | ||
| 3 | 16 | 2 | 3,046 | 0.889 | 0.997 (0.0005) | ||
N opt the optimum number of recombinant inbred lines, h 2opt the optimum heritability on an entry mean basis, B the total budget in million $, l the number of quantitative trait loci, VC the ratio of variance components
The costs for (1) establishing one RIL (C dev), (2) genotyping one RIL (C geno), and (3) testing one field plot (C fp), and the fixed costs for each environment (C env) were 30, 1,000, 15, and 25,000 $, respectively. For a detailed definition of the examined parameters see “Materials and methods”
