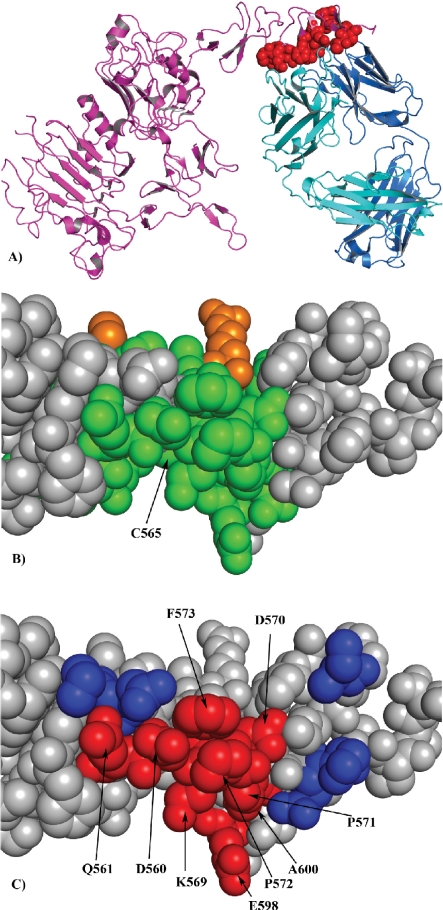
Figure 3.
Comparison of the genuine epitope site on HER2 surface obtained from X-ray crystallography with the epitope site predicted by the EpiSearch method. A) The amino acids at the genuine epitope site on HER-2 surface (red). B) The amino acids predicted by the EpiSearch method present in the highest scoring patch centered on C565 and also present in the input peptide sequences (green), while the residues not present in the input peptide sequences are shown in orange. C) Amino acids correctly predicted and present in the genuine epitope binding site on HER2 surface (red), while the residues present in the epitope site and not predicted are shown in blue.