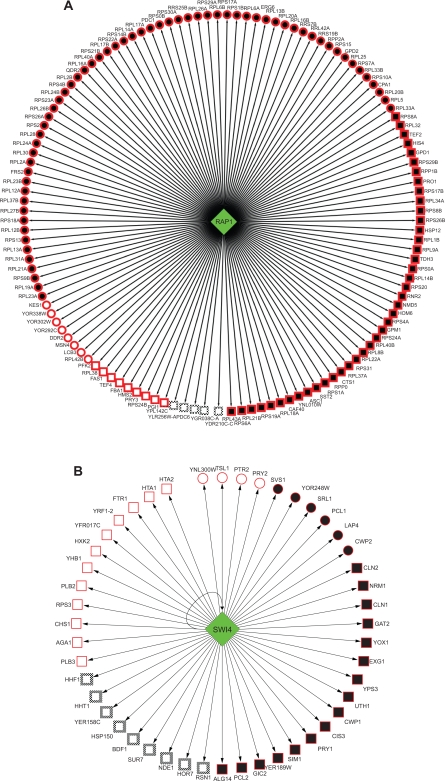
Figure 3.
Comparison with high-quality ChIP-chip data. Oval nodes are for genes identified with stringent P-value cutoffs (Pbt = 0.001, Pet = 0.001), while rectangular nodes are for additional genes identified using optimal relaxed threshold pair by our method. Nodes with red solid border are for relations supported by YEASTRACT, otherwise with black dash border. Solid nodes are for the genes supported by high-quality ChIP-chip data. A) 126 identified target genes of RAP1. 56 additional target genes are identified (rectangular), while 51 (rectangular with red solid border) are supported by YEASTRACT and 34 (solid rectangular) are supported by high-quality ChIP-chip data. B) We have identified 48 target genes of SWI4 including SWI4 itself. SWI4-SWI4 self-regulation is showed by the arrow pointed back to SWI4 itself in the figure. Among SWI4 and other 37 additional target genes identified using optimal relaxed threshold pair by our method (rectangular), as many as 28 (rectangular with red solid border) and SWI4 are supported by YEASTRACT; 16 (solid rectangular) and SWI4 are supported by high-quality ChIP-chip data.