Figure 5.
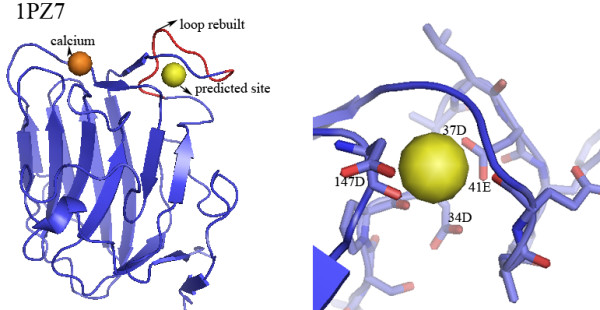
Novel FEATURE predictions. The structure of chicken argin G3 (PDB ID: 1PZ7) and the predicted site. The core structure of 1PZ7 is a beta-sandwich. The top edge of the beta-sandwich consists of four loops, which correspond to a versatile molecular recognition surface in argin. Calcium binding is observed near residues 137-144. Loop 32-40 (residues 32-40, sequence: SPDALDYPA) is unstructured in the original PDB structure. FEATURE predicts a binding site near loop 32-40 when the structure of this region was built using modeling methods. The predicted site is 13.52 Å away from the experimentally observed site; therefore it is a novel site. The close-up view (red for oxygen atoms) shows the close associations between the predicted site and two residues 34D, 37D, 41E and 147D. The overall fold of the two binding sites shares high homology with the C2 calcium-binding domain. The loops in the C2 domain often coordinate 2-3 calcium ions. This suggests our prediction of the second calcium-binding site is reasonable.
