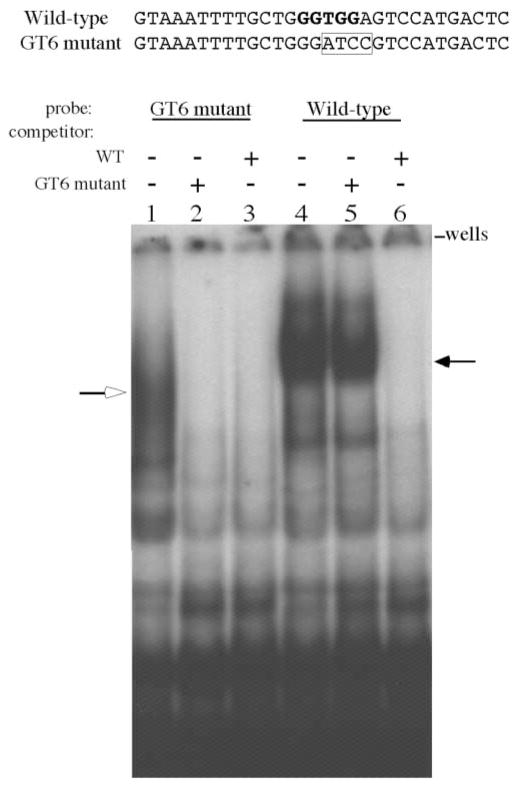
Figure 2.
Electrophoretic mobility shift assay (EMSA) of MEL nuclear extracts with probes derived from the GT6 motif of the HS3 core element. Sequences of the wild-type GT6 motif (WT) and mutant GT6 motif (GT6m) are shown above the autroradiogram. The WT sequence has the GT motif shown in bold and the GT6m sequence shows the four nucleotide substitutions (boxed). A 32P-labeled double stranded oligonucleotide encompassing the WT or mutant GT6 motif was used in the electrophoretic mobility shift assay. Nuclear extracts were prepared from MEL cells as described in the Materials and Methods. Lanes 1–3 used the GT6m probe: lane 1 no competitor; lane 2, competed with unlabeled GT6m probe; lane 3, competed with unlabeled WT probe. Unlabeled competitor DNA was added at 200-fold molar excess. Notice the binding of a complex designated by an open arrow that was effectively competed by both the GT6m and WT probes. Lanes 4–6 used the WT sequence as the probe; lane 4, no competitor; lane 5, competed with unlabeled GT6m probe; lane 6, competed with unlabeled WT probe. A slow migrating complex (filled arrow) is shown in lane 4 and in lane 5 where unlabeled GT6m was used as a competitor. Unlabeled WT probe easily competed for this complex as seen in lane 6.