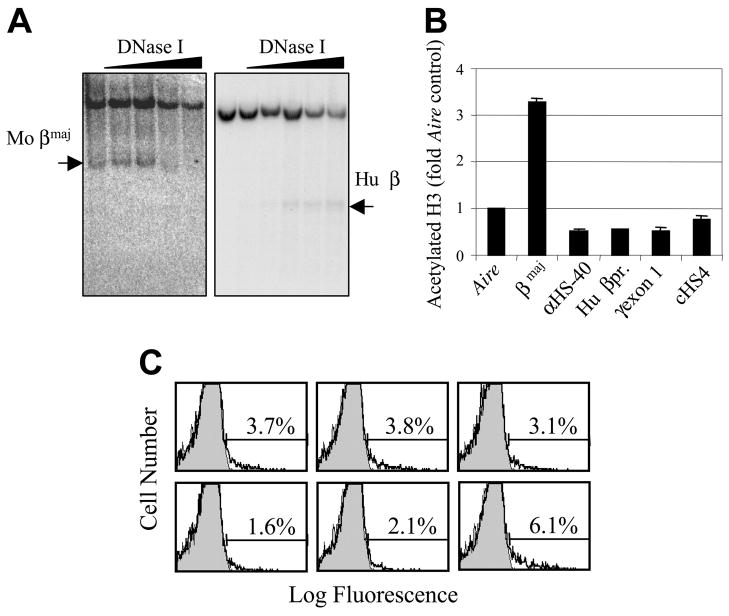
Figure 7. Nature of transgene silencing.
(A) DNase I hypersensitivity assay. Nuclei from splenocytes containing the cHS4/HS-40/βpr/γ gene transgene were treated with increasing amounts of DNase I, and then purified DNA was digested completely with EcoRI (right) or ScaI (left), Southern blotted, and hybridized with a probe for 3′ γ-globin (right) or a probe for the mouse βmaj gene (left). The arrow indicates the subband corresponding to specific digestion at the human β gene promoter (Hu β) and the mouse βmaj gene (Mo βmaj). (B) Chromatin immunoprecipitation (ChIP) assay. Splenocytes containing the cHS4/HS-40/βpr/γ gene transgene were cross-linked with formaldehyde and sonicated, and the soluble chromatin was immunoprecipitated with an antibody against acetylated histone H3. The immunoprecipitated DNA was purified and quantified by real-time PCR with the use of primers specific for the indicated portions of the transgene, the endogenous mouse βmaj gene (as a positive control), and the endogenous mouse Aire gene (as an unexpressed negative control). All data are expressed as fold differences compared with the mouse Aire gene. Error bars indicate standard deviation (SD). (C) Immunofluorescent analysis of γ-globin transgene expression. Red blood cells (RBCs) from mice containing the HS4/HS-40/βpr/γ gene were fixed, permeabilized, stained with a phycoerythrin–conjugated anti-HbF antibody, and analyzed by flow cytometry. Profiles for transgenic animals (unfilled histogram with bold line) are overlaid on a profile from a nontransgenic control (filled histogram, thin line). The percentage of positive cells is presented above the indicated gates.