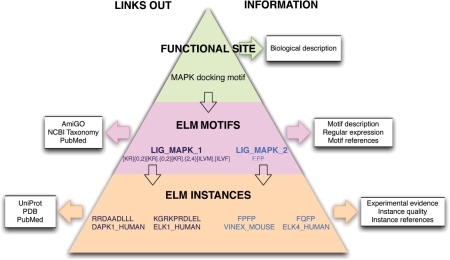
Figure 1.
The ELM Resource hierarchy represented as a pyramid. ‘Functional Site’ provides a general description of the biology, for example, MAP Kinases have a docking motif in their substrates. There are more than one class of MAPK docking motifs and ELM currently provides two ‘ELM Motif’ entries. These contain the motif regular expression and are annotated with more specific information as well as linking out to remote resources including PubMed, NCBI Taxonomy and GO. At the base of the pyramid are the ‘ELM Instances’ that belong to a given ‘ELM Motif’ entry. The instances are annotated with information about experimental methods and instance quality and link to external resources including UniProt, PDB and PubMed.