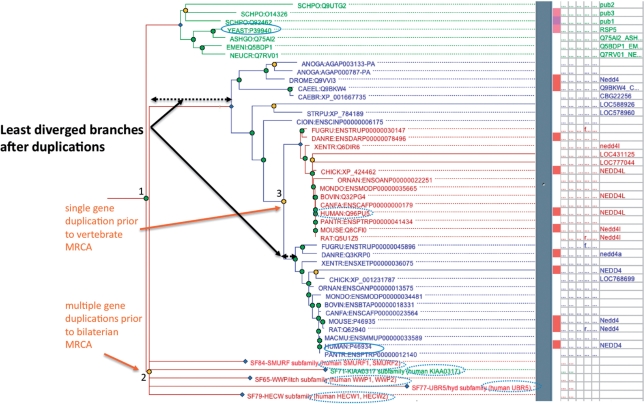
Figure 2.
Example of human orthologs and LDO of the yeast RSP5 gene, identified using a phylogenetic tree. The figure shows part of the tree for PTHR11254 (HECT domain ubiquitin–protein ligase family), tracing the evolutionary relationship between RSP5 and its orthologs in humans, particularly its LDO, NEDD4. Orange nodes represent gene duplication events, green nodes represent speciation events, blue nodes represent subfamily nodes; in this figure blue nodes represent genes present in the bilaterian common ancestor that went on to found subfamilies. The solid outline ovals indicate the LDO pair in human and yeast, RSP5 and NEDD4 respectively. RSP5 has an additional nine orthologs in humans (dashed-outline ovals), but these have diverged to a greater degree than NEDD4. Conversely, 10 human genes have RSP5 as the ortholog, but only NEDD4 has RSP5 as the LDO. The LDO is identified by starting with the MRCA, and following the branch with the shortest length (least sequence divergence) after each gene duplication event. In this example, the MRCA is the speciation event that separated NEDD4 from RSP5 (labeled ‘1’), and there are at least two gene duplication events in the NEDD4 lineage: one at the base of the bilaterians representing multiple events that occurred in relatively rapid succession (labeled ‘2’) to create six genes in total and one at the base of the vertebrates (labeled ‘3’) to create the ancestors of NEDD4 and NEDD4L.