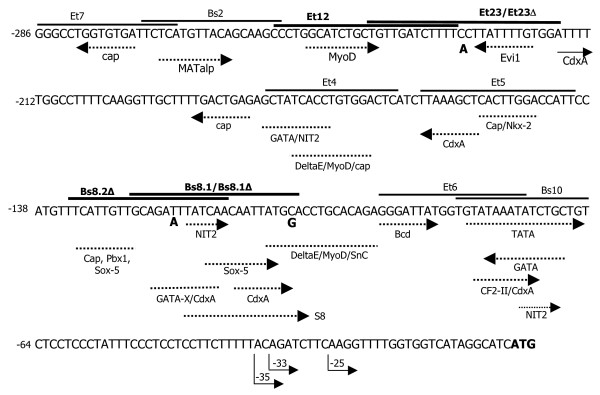
Figure 1.
PbGP43 5' proximal flanking region from Pb339 between -286 and -1 showing the positions of oligonucleotides tested by EMSA and putative transcription motifs. ATG start codon is bolded. Oligonucleotides that formed EMSA specific bands are indicated with bolded names. The three substitutions that occur in isolates from PS2 phylogenetic group are indicated at -104, -130, and -230, as well as the three transcription start sites mapped in four different isolates [16]. The positions of some putative transcription motifs detected with the TFsearch program http://www.cbrc.jp/research/db/TFSEARCH.html are indicated with the correspondent transcription factor. When the -120 (A) and -103 (G) mutations are considered, we point out disappearance of a GATA-X binding site and introduction of a C/EBP motif in Bs8.2Δ.