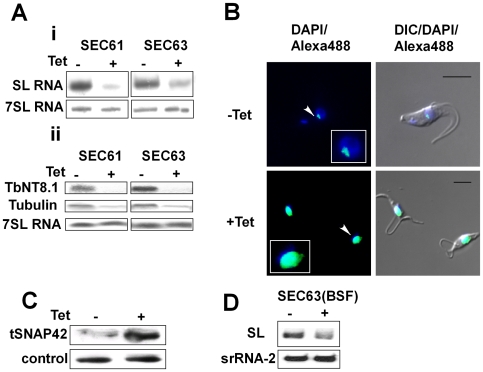
Figure 5. Silencing of SEC61 and SEC63 induces SLS.
A. Northern blot analysis of SEC63 and SEC61 RNAs. RNA was prepared from uninduced (-Tet) and silenced cells 3 days after induction (+Tet) and subjected to Northern analysis with radio-labeled probes. i. Small RNAs; ii. mRNAs. The transcripts examined are indicated. B. Immunofluorescence monitoring the changes in amount and localization of tSNAP42 during SEC63 silencing. Uninduced cells (-Tet) and cells induced (+Tet) for 48 hours were fixed with 4% (v/v) formaldehyde for 25 min, incubated with tSNAP42 antibodies and detected as described in Materials and Methods. The nucleus was stained with DAPI. DIC- differential interference contrast. The localization of tSNAP42 is indicated with arrows. Scale bars, 5 µm. Framed region shows enlargement of the nuclear area. C. Changes in the level of tSNAP42 in cells carrying SEC63 silencing construct. Western blot analysis using anti-tSNAP42 antibodies. Nuclei were extracted from uninduced cells (-Tet) and cells induced (+Tet) for 48 hours, as previously described [23]. The extracts were separated on 10% SDS-PAGE and reacted with anti tSNAP42 antibodies. Rabbit anti PTB1 antiserum was used as a loading control. D. Northern blot analysis of RNA from bloodstream trypanosomes silenced for SEC63. RNA was prepared from uninduced (-Tet) and silenced cells 3 days after induction (+Tet) and subjected to Northern analysis with radio-labeled probes. srRNA-2 probe was used for loading control.