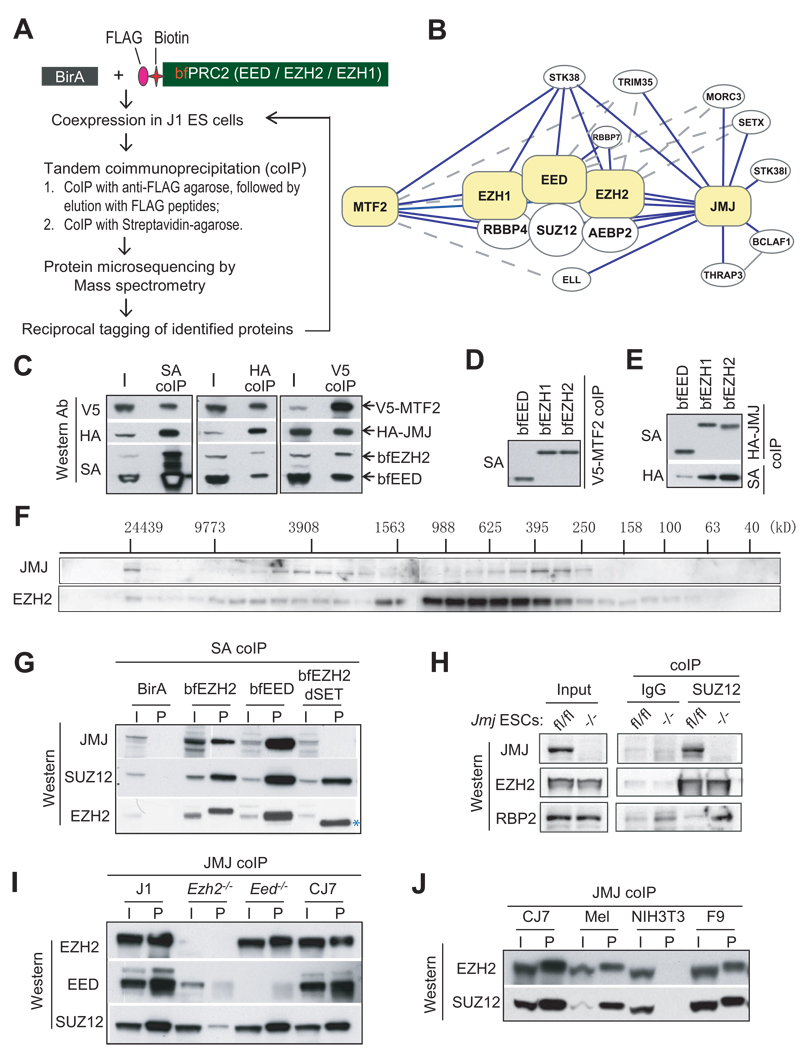
Figure 1. JMJ interacts with the PRC2 complex.
(A) The scheme to identify PRC2 interacting proteins.
(B) A PRC2 interactome. Proteins highlighted in yellow are tagged baits for affinity purification. Solid and dotted lines indicate interactions identified by multiple or few replicates, respectively. The interaction between BCLAF1 and THRAP3 was reported previously (Bracken et al., 2008).
(C) – (E) Coimmunoprecipitation (coIP) analysis in 293T cells. V5-tagged MTF2, HA-tagged JMJ, bfEED and bfEZH2 were coexpressed (C). V5-MTF2 (D) or HA-JMJ (E) was coexpressed with bfEED, bfEZH1, or bfEZH2 as indicated.
(F) Gel filtration analysis of nuclear extracts from CJ7 ESCs. Migration of molecular markers is indicated above the panels, and the antibodies for Western blotting are shown on the left.
(G) Streptavidin (SA) mediated coIP in ESCs expressing birA alone, bfEED, bfEZH2 or mutant EZH2 with SET domain deleted (bfEZH2dSET). bfEZH2dSET is indicated by *.
(H) Anti-SUZ12 coIP in Jmfl/fl and Jmj−/− ESCs.
(I) – (J) Anti-JMJ antibody-mediated coIP in ESCs and non-pluripotent cells. In (C) – (J), “I” indicates inputs and “P” indicates coIP fractions. About 5% of nuclear extracts used for coIP were loaded as the inputs.