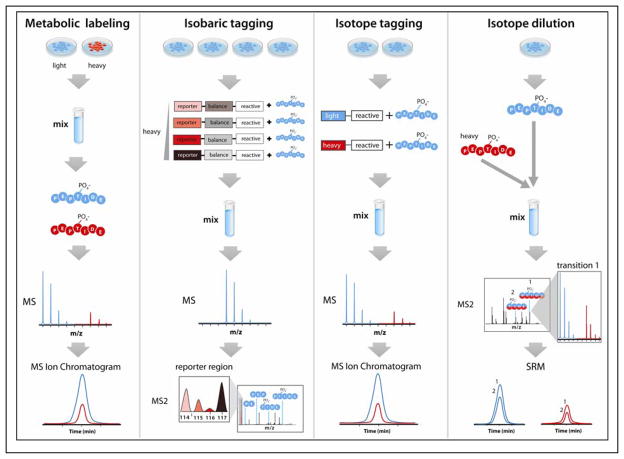
Figure 4. Isotope labeling strategies for phosphopeptide quantitation.
Metabolic labeling introduces heavy isotopes into proteins synthesized in living cells, using heavy isotope-containing amino acids (SILAC) or nutrients (15N or 13C). Peptides analyzed by MS exhibit an m/z difference between light- and heavy-labeled peptides, allowing for relative quantitation by monitoring extracted ion chromatograms of eluting peptides. Isobaric tagging strategies (e.g., iTRAQ and TMT) label peptides from different samples after protein digestion is performed. As co-eluting peptides modified with tags having the same nominal mass are isolated and fragmented, the MS2 scan provides assessment of relative abundance through analysis of the intensity of low m/z reporters. Isotope tagging strategies label digested peptides with heavy and light reactive tags (e.g., ICAT), which allow for determining relative abundance from extracted ion chromatograms. Absolute quantitation (AQUA) of phosphopeptides can be achieved using isotope dilution, in which a known amount of a synthetic heavy isotope-labeled phosphopeptide is spiked into a sample after enzymatic digestion is performed to produce the corresponding endogenous phosphopeptide of interest. Quantitation is achieved by selected reaction monitoring (SRM), typically performed on a triple quadrupole mass spectrometer. For all strategies, the step in the workflow which incorporates heavy isotopes is indicated, with blue representing samples with naturally occurring light isotopes and red representing samples containing stable heavy isotopes. Figure panels adapted with permission from the following references (96, 126).