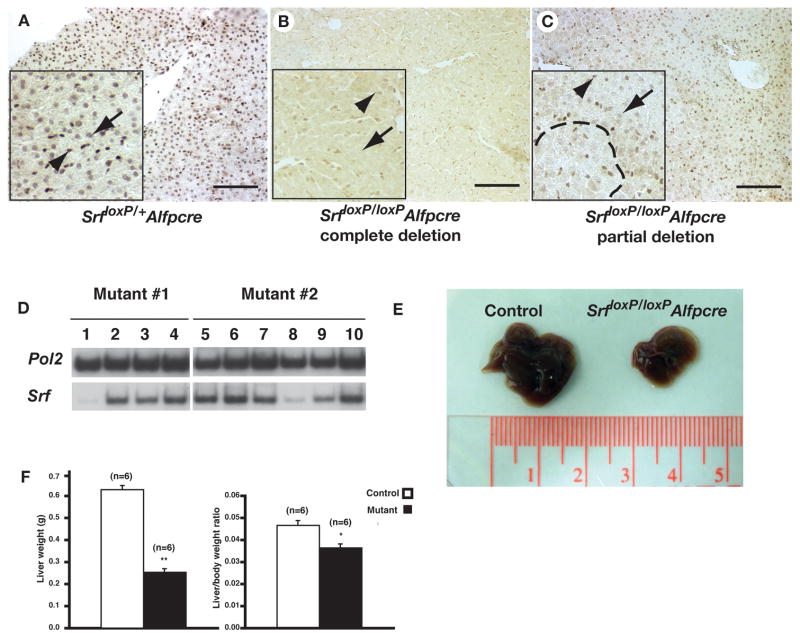
Figure 3. Efficiency of SRF depletion is variable in SrfloxP/loxPAlfpcre livers.
A–C) Immunohistochemistry detecting SRF protein in livers of Srf loxP/+Alfp-Cre (A) and Srf loxP/loxPAlfp-Cre (B, C) mice. Inset shows high-resolution images with hepatocytes indicated by an arrow and endothelial cells by an arrowhead. Dashed line demarcates an area of SRF negative hepatocytes in C. Scale bar = 100μM. D) RT-PCR analysis on multiple liver fragments (numbered) from two Srf loxP/loxPAlfp-Cre mice (mutant #1 and #2) to detect Srf mRNA. Note reduced Srf mRNA levels in samples 1 and 8. Amplification using primers to detect RNA Polymerase II (Pol2) was used to normalize. E) Micrographs comparing the size of typical livers isolated from control (left) and Srf loxP/loxPAlfp-Cre (right) male mice. F) Bar graphs showing the weight of livers (left) and relative body weight to liver ratio (right) in Srf loxP/+Alfp-Cre (white) and Srf loxP/loxPAlfp-Cre (black) mice. Student’s t-test, * p≤0.005, ** p≤0.001.