Figure 6.
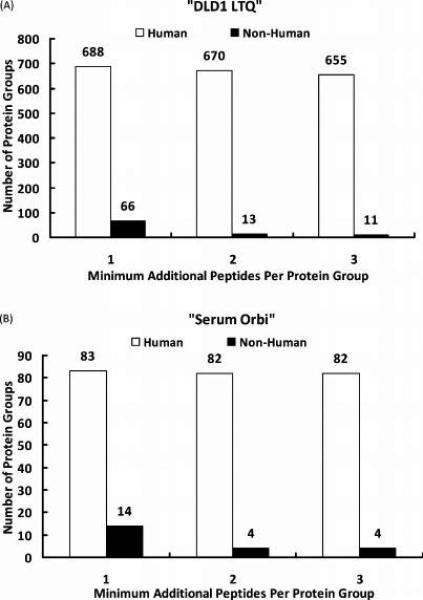
Reduction of orthologous protein identifications in a multispecies database search. Two different human samples (“DLD1 LTQ” and “Serum Orbi”) were matched to the Swiss-Prot multispecies database (version 56.2) using MyriMatch. Protein groups (containing indistinguishable proteins) were assembled from peptide identifications using IDPicker. Three different settings were used for the “Minimum additional peptides per protein group” filter in the assembly process. At each setting, the total numbers of human and nonhuman protein groups were enumerated and plotted for both “DLD1 LTQ” (A) and “Serum Orbi” (B) samples. Setting the filter to 2 dramatically reduced the number of nonhuman (orthologous) protein identifications from a multispecies database search without significantly affecting the number of human (paralogous) protein identifications.
