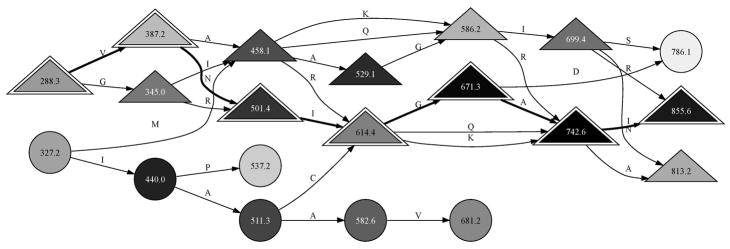
Figure 1.
A spectrum may be viewed as a graph for tag inference. This graph represents the MS/MS observed for the peptide DAGTIAGLNVLR. Each major peak in the spectrum is shown as a node, with the most intense peaks given the darkest shading. When peaks are separated by the mass of an amino acid, they are joined by an edge labeled with the amino acid symbol. Complemented peaks are denoted by triangles. The 3-letter sequence tags for this spectrum are sequences of four connected nodes. DirecTag was configured to retain the top 100 peaks for each spectrum, yielding graphs with far more possible tags than shown here. The true sequence of this peptide can be reconstructed from a path of ions stretching from 288.3 to 855.6 m/z (indicated by outlined triangles). It is reversed with respect to m/z order because the fragments are y ions.