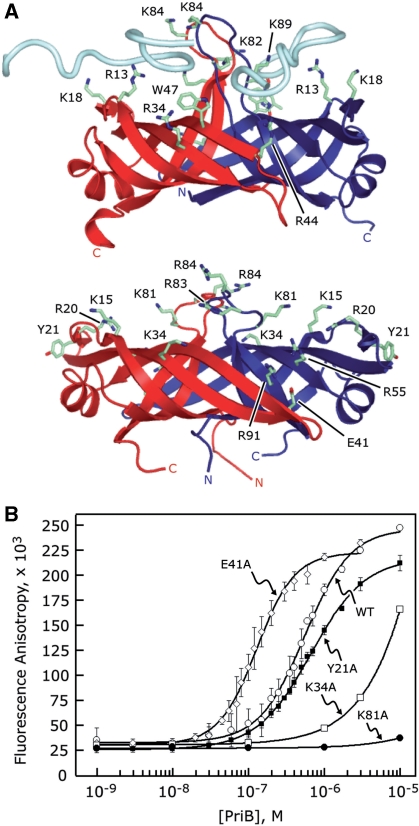
Figure 4.
Identification of PriB’s binding site for ssDNA. (A) Ribbon diagrams of the crystal structures of E. coli PriB bound to ssDNA (top) and apo N. gonorrhoeae PriB (bottom) oriented and colored as in Figure 1. E. coli PriB amino acid residues that are important for binding ssDNA and the analogous residues of N. gonorrhoeae PriB are rendered as sticks and colored according to atom type with carbon atoms colored green, oxygen atoms colored red and nitrogen atoms colored blue. (B) Wild-type PriB (open circles), Y21A (closed squares), K34A (open squares), E41A (open diamonds) and K81A (closed circles) were serially diluted and incubated with a fluorescein-conjugated 36-base ssDNA oligonucleotide as described in the ‘Materials and Methods’ section. Measurements are reported in triplicate and error bars represent 1 SD of the mean.