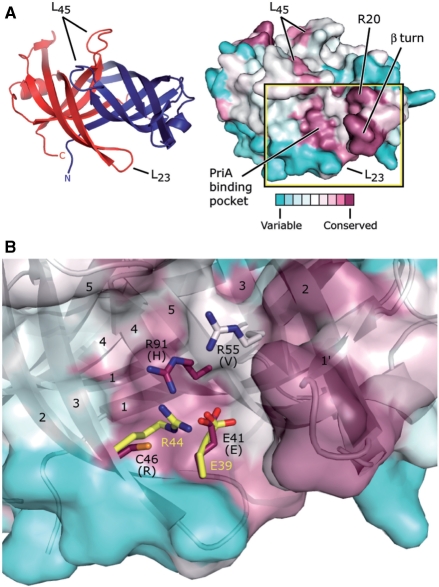
Figure 5.
Potential PriA-binding site on PriB. (A) Ribbon diagram of the crystal structure of N. gonorrhoeae PriB (left) showing frame of reference for the solvent-accessible surface view (right). The ribbon diagram is colored as in Figure 1C and the surface view is colored according to amino acid sequence variability as in Figure 1A. The putative PriA-binding pocket constitutes one of the two main conserved surface patches, and the β turn and amino acid residue R20 constitute the other. (B) Expanded, partially-transparent view of the solvent-accessible surface of PriB inscribed by the box in (A). A ribbon diagram of the crystal structure of PriB underlies the surface view. Individual β strands are numbered and amino acid residues located near residue E41 are labeled and rendered as sticks. Carbon atoms are colored according to amino acid sequence variability as in Figure 1A, oxygen atoms are colored red, nitrogen atoms are colored blue and sulfur atoms are colored orange. For each N. gonorrhoeae PriB residue rendered as sticks, the amino acid residue that corresponds to the PriB consensus sequence is indicated in parentheses. E. coli PriB residues E39 and R44 are superposed with the analogous residues of N. gonorrhoeae PriB and are rendered as sticks. The E. coli PriB residues are colored according to atom type: carbon atoms are colored yellow, oxygen atoms are colored red and nitrogen atoms are colored blue.