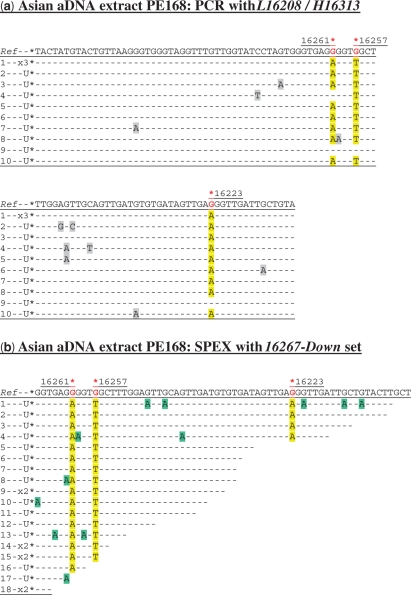
Figure 1.
Cloned sequence data following (a) PCR and (b) SPEX amplification from the Asian aDNA extract PE168. U = a unique sequence found in only one clone from that particular amplification reaction; X1, X2, etc. = identical sequences independently cloned (once, twice, etc). The L16208/H16313 PCR primer pair was used (Supplementary Figure S1a and Table S1). The 16267-Down SPEX primer set (Supplementary Table S1) was used in the amplification strategy described in Supplementary Figure S1a. In relation to the rCRS, hg N9a SNPs are C16223T, C16257A and C16261T. As the 16267-Down SPEX primer set targets the light mtDNA strand, hg N9a-specific SNPs are highlighted by a red asterisk above the reverse complement of the reference sequence (Ref). N9a-specific SNPs in PCR- and SPEX-generated sequence data are highlighted in yellow. In this and subsequent PCR-derived sequence data, base changes at non-diagnostic nucleotide positions—arising from sequence-modifying DNA damage and PCR-generated sequence artefacts—are highlighted in grey. In this and subsequent SPEX-derived sequence data, G > A transition base changes (arising from C > U-type miscoding lesion sequence-modifying DNA damage on template strands) are highlighted in green, with any other non-authentic base changes highlighted in grey. The final 3′-most positions of SPEX amplicons (i.e. the position immediately 5′ to the polyC-tail) do not necessarily reflect underlying template strand sequences, due to potential polymerase-mediated ‘non-templated’ nucleotide addition (NTNA) following SPEX (17). The 3′-most position is therefore not presented, or analysed, in SPEX-generated sequence data.