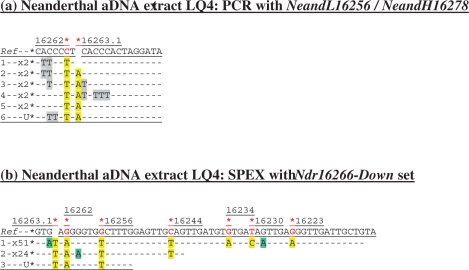
Figure 2.
Cloned sequence data following (a) PCR, and (b) SPEX amplification from the Neanderthal LQ4 aDNA extract. The NeandL16256/NeandH16278 PCR primer pair was used (Supplementary Figure S1b and Table S1). The Ndr16266-Down SPEX primer set (Supplementary Table S1) was used in the amplification strategy described in Supplementary Figure S1b. Nucleotide positions of expected Neanderthal-specific SNPs are each highlighted by a red asterisk above: (a) the reference sequence (Ref); or (b) the reverse complement of the reference sequence (Ref). Presumed Neanderthal-specific SNPs in PCR- and SPEX-generated sequence data are highlighted in yellow. Given above-background levels of C > U-derived G > A transitions in SPEX data, an absence of np 16228 SNPs in published Neanderthal data, and a depth-of-coverage of just one generated for this site by SPEX, the G > A transition base change here is assumed to be due to miscoding lesion DNA damage.