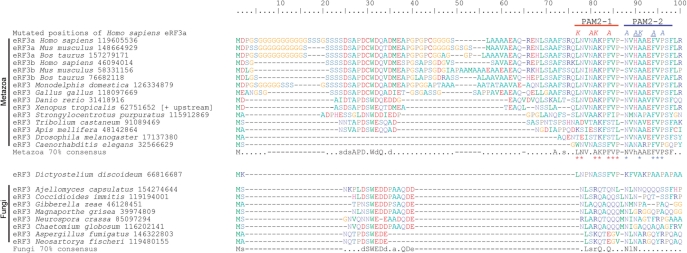
Figure 4.
Consensus and example sequence alignment of the extreme N-terminus of eRF3. Organism names are followed by NCBI GI numbers. The sequence from Xenopus tropicalis was extended at the N-terminus by 15 amino acids, relative to the predicted protein product in Genbank following the discovery of an alternate upstream start codon (mRNA GI number 62751651). The location of the PAM2-1 and PAM2-2 adjacent motifs are indicated above the alignment. Seventy percent conservation consensus sequences were calculated using the Python script ConsensusFinder (GCA). Uppercase letters indicate amino acids conserved in >70% of all examined sequences, and lowercase letters indicate a common amino acid substitution group conserved in >70% of the sequences. A ‘.’ denotes a position that is not conserved in sequence, and gaps are denoted by ‘-’. Asterisks below the Metazoa consensus sequence show residues that match the PAM2 motif, as represented in Pfam. The sites that were mutated are indicated above the human sequence, with the letters indicating the amino acids present in the mutant forms. Red mutated postions: the PAM2-1 KAKA mutant; Blue positions: the PAM2-2 AAKAA mutant; Blue underlined positions: the PAM2-2 AKA mutant.